Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2810-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1783 | 6.134e-03 | 1.655e-03 | 3.932e-02 |
|---|
| IPC-NIBC-129 | 0.2282 | 3.700e-02 | 2.838e-02 | 1.799e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2700 | 1.836e-02 | 1.243e-02 | 2.092e-01 |
|---|
| Loi_GPL570 | 0.2966 | 1.480e-01 | 1.795e-01 | 4.947e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2022 | 7.400e-03 | 3.278e-03 | 9.323e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.4059 | 5.037e-01 | 4.298e-01 | 9.617e-01 |
|---|
| Schmidt | 0.1804 | 2.303e-02 | 2.452e-02 | 1.514e-01 |
|---|
| Sotiriou | 0.2755 | 6.135e-02 | 2.082e-02 | 4.186e-01 |
|---|
| Wang | 0.1633 | 8.961e-02 | 7.338e-02 | 2.785e-01 |
|---|
| Zhang | 0.0965 | 1.781e-03 | 7.430e-04 | 1.109e-02 |
|---|
Expression data for subnetwork 2810-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
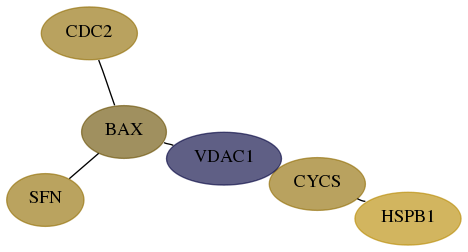
Score for each gene in subnetwork 2810-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| hspb1 |   | 2 | 53 | 115 | 105 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| cycs |   | 2 | 53 | 115 | 105 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | 0.272 | 0.157 | 0.077 | 0.190 | 0.135 |
|---|
| sfn |   | 4 | 27 | 1 | 5 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 |
|---|
| vdac1 |   | 1 | 90 | 115 | 121 | -0.010 | 0.132 | 0.084 | 0.199 | 0.145 | 0.273 | -0.097 | 0.171 | -0.047 | -0.063 |
|---|
| bax |   | 3 | 39 | 1 | 10 | 0.029 | 0.185 | 0.233 | 0.192 | 0.088 | 0.172 | 0.175 | 0.019 | -0.094 | 0.041 |
|---|
GO Enrichment output for subnetwork 2810-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.162E-08 | 4.973E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 2.447E-08 | 2.814E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 3.28E-08 | 2.515E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.152E-06 | 6.626E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.613E-06 | 7.42E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.53E-06 | 1.353E-03 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.941E-06 | 1.623E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 6.586E-06 | 1.894E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.551E-05 | 3.963E-03 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.137E-05 | 4.915E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.137E-05 | 4.468E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.559E-11 | 3.81E-08 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.912E-11 | 2.335E-08 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 2.638E-11 | 2.149E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.135E-09 | 6.93E-07 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.234E-09 | 6.028E-07 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.944E-09 | 7.916E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.496E-08 | 5.22E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.924E-08 | 8.928E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.385E-08 | 9.187E-06 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.447E-08 | 1.086E-05 |
|---|
| nucleus organization | GO:0006997 |  | 2.165E-07 | 4.808E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.097E-11 | 7.451E-08 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 3.564E-11 | 4.288E-08 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 4.983E-11 | 3.996E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.858E-09 | 1.118E-06 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.028E-09 | 9.757E-07 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 3.253E-09 | 1.304E-06 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.458E-08 | 8.448E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 4.803E-08 | 1.445E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 5.561E-08 | 1.487E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 7.305E-08 | 1.758E-05 |
|---|
| nucleus organization | GO:0006997 |  | 3.036E-07 | 6.641E-05 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.559E-11 | 3.81E-08 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.912E-11 | 2.335E-08 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 2.638E-11 | 2.149E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.135E-09 | 6.93E-07 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.234E-09 | 6.028E-07 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.944E-09 | 7.916E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.496E-08 | 5.22E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.924E-08 | 8.928E-06 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.385E-08 | 9.187E-06 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.447E-08 | 1.086E-05 |
|---|
| nucleus organization | GO:0006997 |  | 2.165E-07 | 4.808E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.097E-11 | 7.451E-08 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 3.564E-11 | 4.288E-08 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 4.983E-11 | 3.996E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.858E-09 | 1.118E-06 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.028E-09 | 9.757E-07 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 3.253E-09 | 1.304E-06 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.458E-08 | 8.448E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 4.803E-08 | 1.445E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 5.561E-08 | 1.487E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 7.305E-08 | 1.758E-05 |
|---|
| nucleus organization | GO:0006997 |  | 3.036E-07 | 6.641E-05 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.097E-11 | 7.451E-08 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 3.564E-11 | 4.288E-08 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 4.983E-11 | 3.996E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.858E-09 | 1.118E-06 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.028E-09 | 9.757E-07 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 3.253E-09 | 1.304E-06 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.458E-08 | 8.448E-06 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 4.803E-08 | 1.445E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 5.561E-08 | 1.487E-05 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 7.305E-08 | 1.758E-05 |
|---|
| nucleus organization | GO:0006997 |  | 3.036E-07 | 6.641E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.162E-08 | 4.973E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 2.447E-08 | 2.814E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 3.28E-08 | 2.515E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.152E-06 | 6.626E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.613E-06 | 7.42E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.53E-06 | 1.353E-03 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.941E-06 | 1.623E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 6.586E-06 | 1.894E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.551E-05 | 3.963E-03 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.137E-05 | 4.915E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.137E-05 | 4.468E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.162E-08 | 4.973E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 2.447E-08 | 2.814E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 3.28E-08 | 2.515E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.152E-06 | 6.626E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.613E-06 | 7.42E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.53E-06 | 1.353E-03 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.941E-06 | 1.623E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 6.586E-06 | 1.894E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.551E-05 | 3.963E-03 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.137E-05 | 4.915E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.137E-05 | 4.468E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.162E-08 | 4.973E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 2.447E-08 | 2.814E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 3.28E-08 | 2.515E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.152E-06 | 6.626E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.613E-06 | 7.42E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.53E-06 | 1.353E-03 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.941E-06 | 1.623E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 6.586E-06 | 1.894E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.551E-05 | 3.963E-03 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.137E-05 | 4.915E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.137E-05 | 4.468E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.162E-08 | 4.973E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 2.447E-08 | 2.814E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 3.28E-08 | 2.515E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.152E-06 | 6.626E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.613E-06 | 7.42E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.53E-06 | 1.353E-03 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.941E-06 | 1.623E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 6.586E-06 | 1.894E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.551E-05 | 3.963E-03 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.137E-05 | 4.915E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.137E-05 | 4.468E-03 |
|---|



