Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1894-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1612 | 1.354e-02 | 4.887e-03 | 7.403e-02 |
|---|
| IPC-NIBC-129 | 0.2243 | 9.883e-02 | 8.676e-02 | 4.282e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2323 | 1.982e-02 | 1.360e-02 | 2.226e-01 |
|---|
| Loi_GPL570 | 0.2188 | 1.404e-01 | 1.710e-01 | 4.779e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2114 | 1.448e-02 | 8.064e-03 | 1.996e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3363 | 6.742e-02 | 3.283e-02 | 3.826e-01 |
|---|
| Schmidt | 0.1845 | 2.005e-02 | 2.142e-02 | 1.345e-01 |
|---|
| Sotiriou | 0.2842 | 5.278e-02 | 1.679e-02 | 3.772e-01 |
|---|
| Wang | 0.1615 | 9.225e-02 | 7.609e-02 | 2.845e-01 |
|---|
| Zhang | 0.2910 | 5.834e-03 | 2.789e-03 | 3.001e-02 |
|---|
Expression data for subnetwork 1894-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
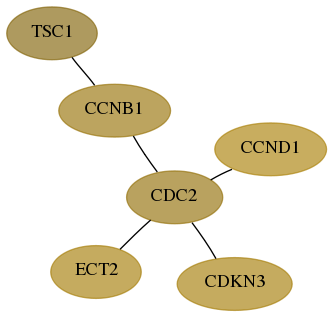
Score for each gene in subnetwork 1894-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| cdkn3 |   | 18 | 9 | 26 | 16 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.323 | 0.186 | 0.198 | 0.134 | 0.271 |
|---|
| tsc1 |   | 31 | 5 | 26 | 15 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 |
|---|
| ect2 |   | 1 | 90 | 110 | 117 | 0.117 | 0.203 | 0.142 | 0.090 | 0.135 | 0.213 | 0.223 | 0.177 | 0.153 | 0.178 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| ccnb1 |   | 33 | 4 | 68 | 40 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 1894-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.749E-08 | 1.552E-04 |
|---|
| interphase | GO:0051325 |  | 7.408E-08 | 8.519E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.152E-06 | 8.835E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.941E-06 | 1.116E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.354E-06 | 1.083E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.354E-06 | 9.024E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.345E-06 | 1.099E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.514E-06 | 1.01E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.056E-06 | 1.036E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.056E-06 | 9.328E-04 |
|---|
| regulation of Rho protein signal transduction | GO:0035023 |  | 4.248E-06 | 8.882E-04 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.811E-08 | 9.31E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.749E-07 | 7.022E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.106E-06 | 9.003E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.234E-06 | 7.537E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.851E-06 | 9.043E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.889E-06 | 7.692E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.975E-06 | 6.891E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.246E-06 | 6.86E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.246E-06 | 6.097E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 2.752E-06 | 6.724E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.974E-06 | 6.605E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.51E-08 | 1.085E-04 |
|---|
| interphase | GO:0051325 |  | 5.114E-08 | 6.152E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.266E-07 | 5.827E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.397E-06 | 8.403E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.454E-06 | 6.998E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.454E-06 | 5.832E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.387E-06 | 8.204E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.495E-06 | 7.503E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.838E-06 | 7.587E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.838E-06 | 6.828E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 3.477E-06 | 7.605E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.811E-08 | 9.31E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.749E-07 | 7.022E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.106E-06 | 9.003E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.234E-06 | 7.537E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.851E-06 | 9.043E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.889E-06 | 7.692E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.975E-06 | 6.891E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.246E-06 | 6.86E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.246E-06 | 6.097E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 2.752E-06 | 6.724E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.974E-06 | 6.605E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.51E-08 | 1.085E-04 |
|---|
| interphase | GO:0051325 |  | 5.114E-08 | 6.152E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.266E-07 | 5.827E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.397E-06 | 8.403E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.454E-06 | 6.998E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.454E-06 | 5.832E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.387E-06 | 8.204E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.495E-06 | 7.503E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.838E-06 | 7.587E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.838E-06 | 6.828E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 3.477E-06 | 7.605E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.51E-08 | 1.085E-04 |
|---|
| interphase | GO:0051325 |  | 5.114E-08 | 6.152E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.266E-07 | 5.827E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.397E-06 | 8.403E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.454E-06 | 6.998E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.454E-06 | 5.832E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.387E-06 | 8.204E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.495E-06 | 7.503E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.838E-06 | 7.587E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.838E-06 | 6.828E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 3.477E-06 | 7.605E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.749E-08 | 1.552E-04 |
|---|
| interphase | GO:0051325 |  | 7.408E-08 | 8.519E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.152E-06 | 8.835E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.941E-06 | 1.116E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.354E-06 | 1.083E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.354E-06 | 9.024E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.345E-06 | 1.099E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.514E-06 | 1.01E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.056E-06 | 1.036E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.056E-06 | 9.328E-04 |
|---|
| regulation of Rho protein signal transduction | GO:0035023 |  | 4.248E-06 | 8.882E-04 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.749E-08 | 1.552E-04 |
|---|
| interphase | GO:0051325 |  | 7.408E-08 | 8.519E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.152E-06 | 8.835E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.941E-06 | 1.116E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.354E-06 | 1.083E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.354E-06 | 9.024E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.345E-06 | 1.099E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.514E-06 | 1.01E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.056E-06 | 1.036E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.056E-06 | 9.328E-04 |
|---|
| regulation of Rho protein signal transduction | GO:0035023 |  | 4.248E-06 | 8.882E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.749E-08 | 1.552E-04 |
|---|
| interphase | GO:0051325 |  | 7.408E-08 | 8.519E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.152E-06 | 8.835E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.941E-06 | 1.116E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.354E-06 | 1.083E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.354E-06 | 9.024E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.345E-06 | 1.099E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.514E-06 | 1.01E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.056E-06 | 1.036E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.056E-06 | 9.328E-04 |
|---|
| regulation of Rho protein signal transduction | GO:0035023 |  | 4.248E-06 | 8.882E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.749E-08 | 1.552E-04 |
|---|
| interphase | GO:0051325 |  | 7.408E-08 | 8.519E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.152E-06 | 8.835E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.941E-06 | 1.116E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.354E-06 | 1.083E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.354E-06 | 9.024E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.345E-06 | 1.099E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.514E-06 | 1.01E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.056E-06 | 1.036E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.056E-06 | 9.328E-04 |
|---|
| regulation of Rho protein signal transduction | GO:0035023 |  | 4.248E-06 | 8.882E-04 |
|---|



