Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1871-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1215 | 2.297e-02 | 1.002e-02 | 1.117e-01 |
|---|
| IPC-NIBC-129 | 0.2193 | 4.347e-02 | 3.412e-02 | 2.101e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2069 | 2.189e-02 | 1.527e-02 | 2.408e-01 |
|---|
| Loi_GPL570 | 0.2839 | 5.614e-02 | 7.376e-02 | 2.464e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1723 | 6.360e-02 | 5.566e-02 | 6.591e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.2816 | 1.006e-01 | 5.485e-02 | 4.977e-01 |
|---|
| Schmidt | 0.2539 | 2.944e-02 | 3.117e-02 | 1.858e-01 |
|---|
| Sotiriou | 0.2599 | 9.937e-02 | 4.147e-02 | 5.659e-01 |
|---|
| Wang | 0.1749 | 7.348e-02 | 5.716e-02 | 2.408e-01 |
|---|
| Zhang | 0.2543 | 1.468e-02 | 7.820e-03 | 6.382e-02 |
|---|
Expression data for subnetwork 1871-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
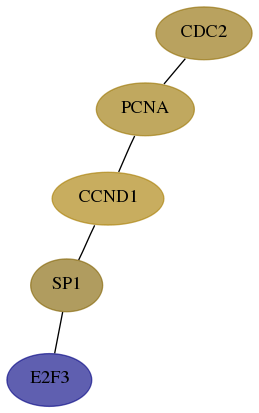
Score for each gene in subnetwork 1871-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| sp1 |   | 31 | 5 | 16 | 8 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 |
|---|
| e2f3 |   | 3 | 39 | 87 | 68 | -0.054 | 0.341 | 0.099 | 0.140 | 0.072 | 0.097 | 0.340 | 0.081 | 0.017 | 0.032 |
|---|
| pcna |   | 39 | 3 | 8 | 3 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 1871-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 4.344E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 7.101E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.08E-05 | 0.02361338 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.519E-05 | 0.02023386 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.519E-05 | 0.01618709 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 4.484E-05 | 0.01718829 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.149E-05 | 0.02020301 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.403E-05 | 0.02128434 |
|---|
| fat cell differentiation | GO:0045444 |  | 8.074E-05 | 0.02063307 |
|---|
| base-excision repair | GO:0006284 |  | 8.074E-05 | 0.01856976 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 8.773E-05 | 0.01834397 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.755E-07 | 6.731E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 7.121E-07 | 8.698E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.068E-06 | 8.697E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.993E-06 | 1.217E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 8.531E-06 | 4.168E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 9.667E-06 | 3.936E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.35E-05 | 4.711E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 1.641E-05 | 5.01E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.307E-05 | 6.262E-03 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 2.307E-05 | 5.636E-03 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 2.491E-05 | 5.533E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.524E-07 | 1.088E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 9.918E-07 | 1.193E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.487E-06 | 1.193E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 1.669E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.188E-05 | 5.716E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.346E-05 | 5.397E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 5.815E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.284E-05 | 6.869E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.965E-05 | 7.926E-03 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 3.211E-05 | 7.726E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.468E-05 | 7.584E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.755E-07 | 6.731E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 7.121E-07 | 8.698E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.068E-06 | 8.697E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.993E-06 | 1.217E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 8.531E-06 | 4.168E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 9.667E-06 | 3.936E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.35E-05 | 4.711E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 1.641E-05 | 5.01E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.307E-05 | 6.262E-03 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 2.307E-05 | 5.636E-03 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 2.491E-05 | 5.533E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.524E-07 | 1.088E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 9.918E-07 | 1.193E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.487E-06 | 1.193E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 1.669E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.188E-05 | 5.716E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.346E-05 | 5.397E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 5.815E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.284E-05 | 6.869E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.965E-05 | 7.926E-03 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 3.211E-05 | 7.726E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.468E-05 | 7.584E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.524E-07 | 1.088E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 9.918E-07 | 1.193E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.487E-06 | 1.193E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.775E-06 | 1.669E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.188E-05 | 5.716E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.346E-05 | 5.397E-03 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.692E-05 | 5.815E-03 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 2.284E-05 | 6.869E-03 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.965E-05 | 7.926E-03 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 3.211E-05 | 7.726E-03 |
|---|
| fat cell differentiation | GO:0045444 |  | 3.468E-05 | 7.584E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 4.344E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 7.101E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.08E-05 | 0.02361338 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.519E-05 | 0.02023386 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.519E-05 | 0.01618709 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 4.484E-05 | 0.01718829 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.149E-05 | 0.02020301 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.403E-05 | 0.02128434 |
|---|
| fat cell differentiation | GO:0045444 |  | 8.074E-05 | 0.02063307 |
|---|
| base-excision repair | GO:0006284 |  | 8.074E-05 | 0.01856976 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 8.773E-05 | 0.01834397 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 4.344E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 7.101E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.08E-05 | 0.02361338 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.519E-05 | 0.02023386 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.519E-05 | 0.01618709 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 4.484E-05 | 0.01718829 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.149E-05 | 0.02020301 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.403E-05 | 0.02128434 |
|---|
| fat cell differentiation | GO:0045444 |  | 8.074E-05 | 0.02063307 |
|---|
| base-excision repair | GO:0006284 |  | 8.074E-05 | 0.01856976 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 8.773E-05 | 0.01834397 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 4.344E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 7.101E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.08E-05 | 0.02361338 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.519E-05 | 0.02023386 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.519E-05 | 0.01618709 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 4.484E-05 | 0.01718829 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.149E-05 | 0.02020301 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.403E-05 | 0.02128434 |
|---|
| fat cell differentiation | GO:0045444 |  | 8.074E-05 | 0.02063307 |
|---|
| base-excision repair | GO:0006284 |  | 8.074E-05 | 0.01856976 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 8.773E-05 | 0.01834397 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.889E-06 | 4.344E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.175E-06 | 7.101E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.08E-05 | 0.02361338 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.519E-05 | 0.02023386 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.519E-05 | 0.01618709 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 4.484E-05 | 0.01718829 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.149E-05 | 0.02020301 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.403E-05 | 0.02128434 |
|---|
| fat cell differentiation | GO:0045444 |  | 8.074E-05 | 0.02063307 |
|---|
| base-excision repair | GO:0006284 |  | 8.074E-05 | 0.01856976 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 8.773E-05 | 0.01834397 |
|---|



