Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 10397-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2181 | 6.640e-03 | 1.845e-03 | 4.192e-02 |
|---|
| IPC-NIBC-129 | 0.1900 | 3.125e-02 | 2.340e-02 | 1.522e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2662 | 3.903e-02 | 3.001e-02 | 3.699e-01 |
|---|
| Loi_GPL570 | 0.3097 | 1.917e-01 | 2.273e-01 | 5.803e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2326 | 1.471e-03 | 3.620e-04 | 9.983e-03 |
|---|
| Pawitan_GPL96-GPL97 | 0.2956 | 2.451e-01 | 1.715e-01 | 7.871e-01 |
|---|
| Schmidt | 0.1602 | 1.276e-02 | 1.377e-02 | 9.045e-02 |
|---|
| Sotiriou | 0.3126 | 3.731e-02 | 1.022e-02 | 2.921e-01 |
|---|
| Wang | 0.1711 | 7.843e-02 | 6.206e-02 | 2.527e-01 |
|---|
| Zhang | 0.1700 | 1.160e-02 | 6.009e-03 | 5.274e-02 |
|---|
Expression data for subnetwork 10397-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
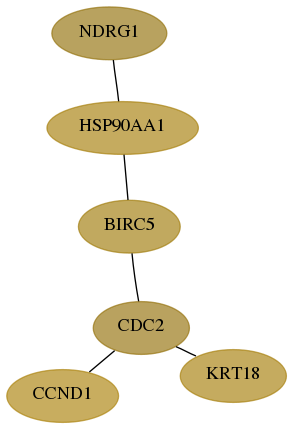
Score for each gene in subnetwork 10397-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| krt18 |   | 13 | 12 | 99 | 67 | 0.112 | -0.078 | 0.147 | 0.150 | 0.161 | 0.083 | -0.099 | 0.118 | 0.086 | -0.114 |
|---|
| hsp90aa1 |   | 1 | 90 | 99 | 108 | 0.116 | 0.030 | 0.164 | 0.076 | 0.119 | 0.169 | 0.069 | 0.102 | 0.052 | -0.011 |
|---|
| ndrg1 |   | 1 | 90 | 99 | 108 | 0.072 | 0.301 | 0.084 | 0.324 | 0.108 | 0.083 | 0.068 | 0.064 | 0.128 | 0.092 |
|---|
| birc5 |   | 3 | 39 | 99 | 83 | 0.101 | 0.216 | 0.190 | 0.134 | 0.223 | 0.202 | 0.236 | 0.241 | 0.124 | 0.205 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
GO Enrichment output for subnetwork 10397-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 0.01029669 |
|---|
| protein refolding | GO:0042026 |  | 7.644E-06 | 8.791E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 7.644E-06 | 5.861E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 7.644E-06 | 4.395E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.921E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.07E-05 | 4.101E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.07E-05 | 3.515E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.07E-05 | 3.075E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.426E-05 | 3.643E-03 |
|---|
| response to metal ion | GO:0010038 |  | 1.709E-05 | 3.93E-03 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 1.832E-05 | 3.831E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.325E-07 | 2.034E-03 |
|---|
| protein refolding | GO:0042026 |  | 2.159E-06 | 2.637E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 3.022E-06 | 2.461E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 3.022E-06 | 1.846E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 3.022E-06 | 1.477E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.022E-06 | 1.23E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.028E-06 | 1.406E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.028E-06 | 1.23E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.028E-06 | 1.093E-03 |
|---|
| mast cell activation | GO:0045576 |  | 4.028E-06 | 9.841E-04 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 4.028E-06 | 8.947E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.366E-06 | 3.287E-03 |
|---|
| protein refolding | GO:0042026 |  | 3.007E-06 | 3.617E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 4.208E-06 | 3.375E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 4.208E-06 | 2.531E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 4.208E-06 | 2.025E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 4.208E-06 | 1.688E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.61E-06 | 1.928E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 5.61E-06 | 1.687E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 5.61E-06 | 1.5E-03 |
|---|
| mast cell activation | GO:0045576 |  | 5.61E-06 | 1.35E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 5.61E-06 | 1.227E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.325E-07 | 2.034E-03 |
|---|
| protein refolding | GO:0042026 |  | 2.159E-06 | 2.637E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 3.022E-06 | 2.461E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 3.022E-06 | 1.846E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 3.022E-06 | 1.477E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.022E-06 | 1.23E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.028E-06 | 1.406E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.028E-06 | 1.23E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.028E-06 | 1.093E-03 |
|---|
| mast cell activation | GO:0045576 |  | 4.028E-06 | 9.841E-04 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 4.028E-06 | 8.947E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.366E-06 | 3.287E-03 |
|---|
| protein refolding | GO:0042026 |  | 3.007E-06 | 3.617E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 4.208E-06 | 3.375E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 4.208E-06 | 2.531E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 4.208E-06 | 2.025E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 4.208E-06 | 1.688E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.61E-06 | 1.928E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 5.61E-06 | 1.687E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 5.61E-06 | 1.5E-03 |
|---|
| mast cell activation | GO:0045576 |  | 5.61E-06 | 1.35E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 5.61E-06 | 1.227E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.366E-06 | 3.287E-03 |
|---|
| protein refolding | GO:0042026 |  | 3.007E-06 | 3.617E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 4.208E-06 | 3.375E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 4.208E-06 | 2.531E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 4.208E-06 | 2.025E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 4.208E-06 | 1.688E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.61E-06 | 1.928E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 5.61E-06 | 1.687E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 5.61E-06 | 1.5E-03 |
|---|
| mast cell activation | GO:0045576 |  | 5.61E-06 | 1.35E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 5.61E-06 | 1.227E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 0.01029669 |
|---|
| protein refolding | GO:0042026 |  | 7.644E-06 | 8.791E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 7.644E-06 | 5.861E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 7.644E-06 | 4.395E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.921E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.07E-05 | 4.101E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.07E-05 | 3.515E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.07E-05 | 3.075E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.426E-05 | 3.643E-03 |
|---|
| response to metal ion | GO:0010038 |  | 1.709E-05 | 3.93E-03 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 1.832E-05 | 3.831E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 0.01029669 |
|---|
| protein refolding | GO:0042026 |  | 7.644E-06 | 8.791E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 7.644E-06 | 5.861E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 7.644E-06 | 4.395E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.921E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.07E-05 | 4.101E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.07E-05 | 3.515E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.07E-05 | 3.075E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.426E-05 | 3.643E-03 |
|---|
| response to metal ion | GO:0010038 |  | 1.709E-05 | 3.93E-03 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 1.832E-05 | 3.831E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 0.01029669 |
|---|
| protein refolding | GO:0042026 |  | 7.644E-06 | 8.791E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 7.644E-06 | 5.861E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 7.644E-06 | 4.395E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.921E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.07E-05 | 4.101E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.07E-05 | 3.515E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.07E-05 | 3.075E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.426E-05 | 3.643E-03 |
|---|
| response to metal ion | GO:0010038 |  | 1.709E-05 | 3.93E-03 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 1.832E-05 | 3.831E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.477E-06 | 0.01029669 |
|---|
| protein refolding | GO:0042026 |  | 7.644E-06 | 8.791E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 7.644E-06 | 5.861E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 7.644E-06 | 4.395E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.07E-05 | 4.921E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.07E-05 | 4.101E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.07E-05 | 3.515E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.07E-05 | 3.075E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.426E-05 | 3.643E-03 |
|---|
| response to metal ion | GO:0010038 |  | 1.709E-05 | 3.93E-03 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 1.832E-05 | 3.831E-03 |
|---|



