Study run-b1
Study informations
127 subnetworks in total page | file
236 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 10153-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1587 | 1.113e-02 | 3.741e-03 | 6.341e-02 |
|---|
| IPC-NIBC-129 | 0.2100 | 2.387e-02 | 1.718e-02 | 1.154e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2417 | 2.633e-02 | 1.896e-02 | 2.779e-01 |
|---|
| Loi_GPL570 | 0.3306 | 5.643e-02 | 7.411e-02 | 2.474e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1605 | 1.592e-02 | 9.145e-03 | 2.200e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3083 | 7.625e-02 | 3.844e-02 | 4.161e-01 |
|---|
| Schmidt | 0.2535 | 1.619e-02 | 1.738e-02 | 1.117e-01 |
|---|
| Sotiriou | 0.2977 | 1.200e-01 | 5.426e-02 | 6.275e-01 |
|---|
| Wang | 0.1413 | 1.298e-01 | 1.164e-01 | 3.626e-01 |
|---|
| Zhang | 0.2798 | 9.356e-03 | 4.726e-03 | 4.426e-02 |
|---|
Expression data for subnetwork 10153-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Subnetwork structure for each dataset
- Desmedt
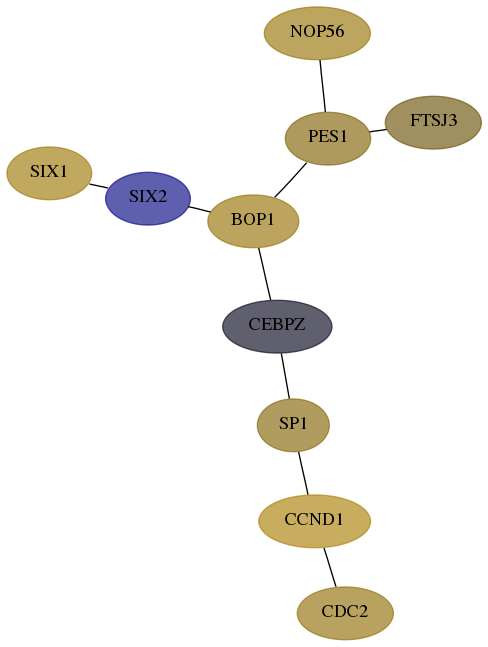
Score for each gene in subnetwork 10153-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang |
|---|
| cebpz |   | 1 | 90 | 74 | 85 | -0.003 | 0.077 | -0.007 | 0.058 | 0.086 | -0.030 | 0.143 | 0.098 | -0.063 | 0.136 |
|---|
| bop1 |   | 1 | 90 | 74 | 85 | 0.083 | 0.081 | 0.080 | 0.106 | 0.101 | 0.220 | 0.141 | 0.040 | -0.109 | 0.080 |
|---|
| pes1 |   | 1 | 90 | 74 | 85 | 0.049 | 0.114 | 0.155 | 0.128 | 0.103 | 0.088 | 0.177 | 0.048 | -0.097 | 0.100 |
|---|
| ftsj3 |   | 1 | 90 | 74 | 85 | 0.029 | 0.177 | 0.189 | 0.136 | 0.118 | 0.150 | 0.132 | 0.105 | 0.068 | -0.052 |
|---|
| six1 |   | 1 | 90 | 74 | 85 | 0.097 | 0.114 | 0.095 | 0.149 | 0.151 | 0.123 | 0.113 | 0.184 | 0.124 | 0.191 |
|---|
| ccnd1 |   | 97 | 2 | 8 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 |
|---|
| sp1 |   | 31 | 5 | 16 | 8 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 |
|---|
| cdc2 |   | 99 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 |
|---|
| six2 |   | 1 | 90 | 74 | 85 | -0.053 | 0.195 | 0.158 | 0.113 | 0.095 | -0.037 | 0.071 | 0.094 | 0.090 | -0.065 |
|---|
| nop56 |   | 1 | 90 | 74 | 85 | 0.084 | 0.176 | 0.209 | 0.287 | 0.111 | 0.219 | 0.116 | 0.086 | 0.088 | 0.111 |
|---|
GO Enrichment output for subnetwork 10153-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| rRNA metabolic process | GO:0016072 |  | 1.01E-08 | 2.323E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.497E-07 | 4.021E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 8.282E-07 | 6.349E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.102E-06 | 6.334E-04 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 2.515E-06 | 1.157E-03 |
|---|
| metanephros development | GO:0001656 |  | 2.783E-06 | 1.067E-03 |
|---|
| ear morphogenesis | GO:0042471 |  | 3.375E-06 | 1.109E-03 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 7.065E-06 | 2.031E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.719E-06 | 2.228E-03 |
|---|
| ear development | GO:0043583 |  | 1.201E-05 | 2.762E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 1.433E-05 | 2.996E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.013E-07 | 4.918E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 4.581E-07 | 5.595E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 6.43E-07 | 5.236E-04 |
|---|
| metanephros development | GO:0001656 |  | 8.718E-07 | 5.325E-04 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 1.479E-06 | 7.229E-04 |
|---|
| ear morphogenesis | GO:0042471 |  | 2.01E-06 | 8.185E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.022E-06 | 1.055E-03 |
|---|
| muscle cell migration | GO:0014812 |  | 3.321E-06 | 1.014E-03 |
|---|
| virus-infected cell apoptosis | GO:0006926 |  | 3.321E-06 | 9.015E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 3.321E-06 | 8.113E-04 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 4.324E-06 | 9.602E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.41E-07 | 5.799E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.706E-07 | 6.865E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 8.117E-07 | 6.51E-04 |
|---|
| metanephros development | GO:0001656 |  | 1.227E-06 | 7.38E-04 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 1.617E-06 | 7.781E-04 |
|---|
| ear morphogenesis | GO:0042471 |  | 2.254E-06 | 9.038E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 4.185E-06 | 1.438E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.185E-06 | 1.259E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.249E-06 | 1.136E-03 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 5.11E-06 | 1.229E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 6.274E-06 | 1.372E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.013E-07 | 4.918E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 4.581E-07 | 5.595E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 6.43E-07 | 5.236E-04 |
|---|
| metanephros development | GO:0001656 |  | 8.718E-07 | 5.325E-04 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 1.479E-06 | 7.229E-04 |
|---|
| ear morphogenesis | GO:0042471 |  | 2.01E-06 | 8.185E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.022E-06 | 1.055E-03 |
|---|
| muscle cell migration | GO:0014812 |  | 3.321E-06 | 1.014E-03 |
|---|
| virus-infected cell apoptosis | GO:0006926 |  | 3.321E-06 | 9.015E-04 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 3.321E-06 | 8.113E-04 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 4.324E-06 | 9.602E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.41E-07 | 5.799E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.706E-07 | 6.865E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 8.117E-07 | 6.51E-04 |
|---|
| metanephros development | GO:0001656 |  | 1.227E-06 | 7.38E-04 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 1.617E-06 | 7.781E-04 |
|---|
| ear morphogenesis | GO:0042471 |  | 2.254E-06 | 9.038E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 4.185E-06 | 1.438E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.185E-06 | 1.259E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.249E-06 | 1.136E-03 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 5.11E-06 | 1.229E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 6.274E-06 | 1.372E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.41E-07 | 5.799E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.706E-07 | 6.865E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 8.117E-07 | 6.51E-04 |
|---|
| metanephros development | GO:0001656 |  | 1.227E-06 | 7.38E-04 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 1.617E-06 | 7.781E-04 |
|---|
| ear morphogenesis | GO:0042471 |  | 2.254E-06 | 9.038E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 4.185E-06 | 1.438E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.185E-06 | 1.259E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.249E-06 | 1.136E-03 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 5.11E-06 | 1.229E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 6.274E-06 | 1.372E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| rRNA metabolic process | GO:0016072 |  | 1.01E-08 | 2.323E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.497E-07 | 4.021E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 8.282E-07 | 6.349E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.102E-06 | 6.334E-04 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 2.515E-06 | 1.157E-03 |
|---|
| metanephros development | GO:0001656 |  | 2.783E-06 | 1.067E-03 |
|---|
| ear morphogenesis | GO:0042471 |  | 3.375E-06 | 1.109E-03 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 7.065E-06 | 2.031E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.719E-06 | 2.228E-03 |
|---|
| ear development | GO:0043583 |  | 1.201E-05 | 2.762E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 1.433E-05 | 2.996E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| rRNA metabolic process | GO:0016072 |  | 1.01E-08 | 2.323E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.497E-07 | 4.021E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 8.282E-07 | 6.349E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.102E-06 | 6.334E-04 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 2.515E-06 | 1.157E-03 |
|---|
| metanephros development | GO:0001656 |  | 2.783E-06 | 1.067E-03 |
|---|
| ear morphogenesis | GO:0042471 |  | 3.375E-06 | 1.109E-03 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 7.065E-06 | 2.031E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.719E-06 | 2.228E-03 |
|---|
| ear development | GO:0043583 |  | 1.201E-05 | 2.762E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 1.433E-05 | 2.996E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| rRNA metabolic process | GO:0016072 |  | 1.01E-08 | 2.323E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.497E-07 | 4.021E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 8.282E-07 | 6.349E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.102E-06 | 6.334E-04 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 2.515E-06 | 1.157E-03 |
|---|
| metanephros development | GO:0001656 |  | 2.783E-06 | 1.067E-03 |
|---|
| ear morphogenesis | GO:0042471 |  | 3.375E-06 | 1.109E-03 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 7.065E-06 | 2.031E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.719E-06 | 2.228E-03 |
|---|
| ear development | GO:0043583 |  | 1.201E-05 | 2.762E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 1.433E-05 | 2.996E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| rRNA metabolic process | GO:0016072 |  | 1.01E-08 | 2.323E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.497E-07 | 4.021E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 8.282E-07 | 6.349E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.102E-06 | 6.334E-04 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 2.515E-06 | 1.157E-03 |
|---|
| metanephros development | GO:0001656 |  | 2.783E-06 | 1.067E-03 |
|---|
| ear morphogenesis | GO:0042471 |  | 3.375E-06 | 1.109E-03 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 7.065E-06 | 2.031E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.719E-06 | 2.228E-03 |
|---|
| ear development | GO:0043583 |  | 1.201E-05 | 2.762E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 1.433E-05 | 2.996E-03 |
|---|



