Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 9054-10-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2665 | 9.700e-03 | 7.236e-03 | 8.510e-02 |
|---|
| IPC-NIBC-129 | 0.2247 | 2.946e-02 | 4.333e-02 | 3.113e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2513 | 8.461e-03 | 1.428e-02 | 2.483e-01 |
|---|
| Loi_GPL570 | 0.3383 | 1.637e-02 | 1.915e-02 | 8.522e-02 |
|---|
| Loi_GPL96-GPL97 | 0.1249 | 1.700e-05 | 1.390e-04 | 1.470e-04 |
|---|
| Parker_GPL1390 | 0.2475 | 5.137e-02 | 6.374e-02 | 3.998e-01 |
|---|
| Parker_GPL887 | 0.0863 | 2.373e-01 | 2.539e-01 | 2.415e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3531 | 6.081e-01 | 5.415e-01 | 9.866e-01 |
|---|
| Schmidt | 0.2411 | 1.550e-01 | 1.959e-01 | 7.238e-01 |
|---|
| Sotiriou | 0.3386 | 7.807e-03 | 5.929e-03 | 1.633e-01 |
|---|
| Van-De-Vijver | 0.2953 | 2.264e-03 | 3.253e-03 | 1.001e-01 |
|---|
| Zhang | 0.1431 | 2.258e-01 | 1.689e-01 | 4.254e-01 |
|---|
| Zhou | 0.3746 | 7.080e-02 | 5.954e-02 | 4.100e-01 |
|---|
Expression data for subnetwork 9054-10-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
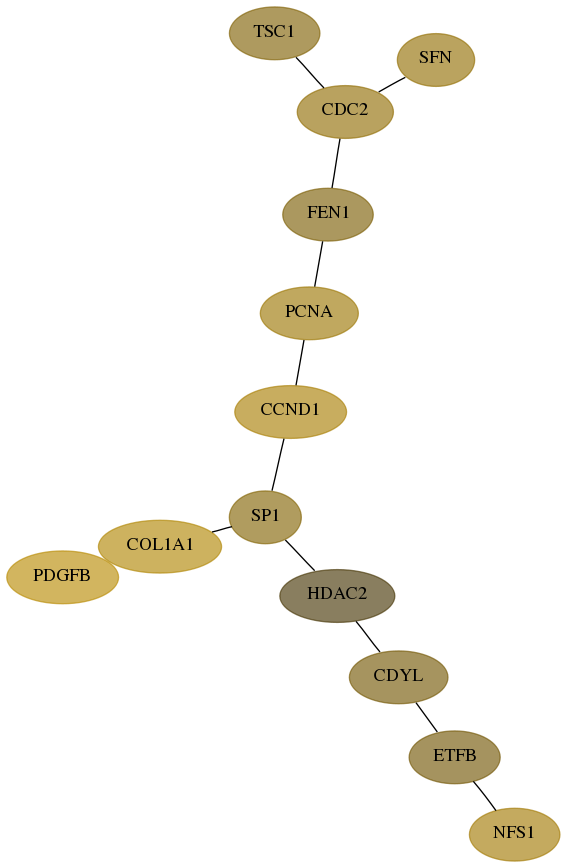
Score for each gene in subnetwork 9054-10-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| etfb |   | 1 | 116 | 175 | 179 | 0.035 | 0.001 | 0.086 | -0.008 | 0.127 | 0.116 | -0.267 | -0.006 | 0.120 | 0.114 | 0.073 | -0.051 | 0.120 |
|---|
| pdgfb |   | 3 | 62 | 95 | 86 | 0.186 | -0.069 | 0.089 | 0.269 | 0.087 | 0.107 | -0.031 | 0.163 | 0.038 | 0.139 | 0.143 | 0.037 | 0.170 |
|---|
| pcna |   | 15 | 11 | 110 | 85 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.119 | 0.104 | 0.256 | 0.151 | 0.059 | 0.201 | 0.123 | 0.233 |
|---|
| fen1 |   | 9 | 22 | 133 | 112 | 0.045 | 0.194 | 0.169 | 0.196 | 0.136 | 0.252 | 0.033 | 0.332 | 0.221 | 0.144 | 0.242 | 0.056 | 0.114 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| sp1 |   | 8 | 26 | 71 | 57 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.033 | -0.370 | 0.202 | 0.284 | 0.227 | 0.304 | 0.091 | -0.068 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| hdac2 |   | 3 | 62 | 146 | 135 | 0.012 | 0.245 | 0.030 | 0.077 | 0.070 | 0.307 | -0.204 | 0.183 | 0.222 | -0.058 | 0.038 | 0.133 | 0.181 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| cdyl |   | 1 | 116 | 175 | 179 | 0.036 | 0.180 | 0.113 | 0.200 | 0.083 | 0.164 | 0.242 | 0.042 | -0.024 | 0.178 | undef | -0.131 | 0.229 |
|---|
| col1a1 |   | 2 | 78 | 142 | 138 | 0.157 | 0.030 | -0.010 | -0.046 | -0.012 | 0.017 | 0.144 | -0.059 | -0.107 | -0.056 | -0.051 | -0.044 | 0.192 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| nfs1 |   | 1 | 116 | 175 | 179 | 0.110 | -0.062 | 0.040 | 0.192 | 0.076 | -0.265 | 0.135 | 0.152 | 0.120 | 0.122 | 0.066 | -0.039 | 0.178 |
|---|
GO Enrichment output for subnetwork 9054-10-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 7.794E-07 | 1.793E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 0.01734556 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 0.01156371 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.508E-05 | 8.673E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 2.261E-05 | 0.0103994 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 9.092E-03 |
|---|
| response to UV | GO:0009411 |  | 2.535E-05 | 8.33E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 2.884E-05 | 8.291E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 8.082E-03 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 3.163E-05 | 7.274E-03 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 3.163E-05 | 6.613E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| metallo-sulfur cluster assembly | GO:0031163 |  | 7.805E-09 | 1.907E-05 |
|---|
| cysteine metabolic process | GO:0006534 |  | 1.365E-08 | 1.668E-05 |
|---|
| skin development | GO:0043588 |  | 4.418E-07 | 3.597E-04 |
|---|
| sulfur amino acid metabolic process | GO:0000096 |  | 6.852E-07 | 4.185E-04 |
|---|
| serine family amino acid metabolic process | GO:0009069 |  | 2.302E-06 | 1.125E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.549E-06 | 2.259E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.549E-06 | 1.937E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.549E-06 | 1.694E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.608E-06 | 1.794E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 8.319E-06 | 2.032E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.319E-06 | 1.848E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 4.853E-07 | 1.168E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.605E-06 | 7.946E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 6.605E-06 | 5.297E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 6.605E-06 | 3.973E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 6.605E-06 | 3.178E-03 |
|---|
| metallo-sulfur cluster assembly | GO:0031163 |  | 6.605E-06 | 2.649E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.526E-06 | 2.93E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 9.903E-06 | 2.978E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 9.903E-06 | 2.647E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 9.903E-06 | 2.383E-03 |
|---|
| response to UV | GO:0009411 |  | 1.152E-05 | 2.519E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| metallo-sulfur cluster assembly | GO:0031163 |  | 7.805E-09 | 1.907E-05 |
|---|
| cysteine metabolic process | GO:0006534 |  | 1.365E-08 | 1.668E-05 |
|---|
| skin development | GO:0043588 |  | 4.418E-07 | 3.597E-04 |
|---|
| sulfur amino acid metabolic process | GO:0000096 |  | 6.852E-07 | 4.185E-04 |
|---|
| serine family amino acid metabolic process | GO:0009069 |  | 2.302E-06 | 1.125E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.549E-06 | 2.259E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.549E-06 | 1.937E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.549E-06 | 1.694E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.608E-06 | 1.794E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 8.319E-06 | 2.032E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.319E-06 | 1.848E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 4.853E-07 | 1.168E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.605E-06 | 7.946E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 6.605E-06 | 5.297E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 6.605E-06 | 3.973E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 6.605E-06 | 3.178E-03 |
|---|
| metallo-sulfur cluster assembly | GO:0031163 |  | 6.605E-06 | 2.649E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.526E-06 | 2.93E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 9.903E-06 | 2.978E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 9.903E-06 | 2.647E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 9.903E-06 | 2.383E-03 |
|---|
| response to UV | GO:0009411 |  | 1.152E-05 | 2.519E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.306E-08 | 8.235E-05 |
|---|
| skin development | GO:0043588 |  | 1.868E-07 | 3.335E-04 |
|---|
| replication fork | GO:0005657 |  | 1.832E-06 | 2.18E-03 |
|---|
| embryonic camera-type eye development | GO:0031076 |  | 6.073E-06 | 5.421E-03 |
|---|
| MutLalpha complex binding | GO:0032405 |  | 6.073E-06 | 4.337E-03 |
|---|
| phosphatidylserine binding | GO:0001786 |  | 6.073E-06 | 3.614E-03 |
|---|
| ribonuclease H activity | GO:0004523 |  | 6.073E-06 | 3.098E-03 |
|---|
| endodeoxyribonuclease activity. producing 5'-phosphomonoesters | GO:0016888 |  | 6.073E-06 | 2.711E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 6.073E-06 | 2.409E-03 |
|---|
| metallo-sulfur cluster assembly | GO:0031163 |  | 6.073E-06 | 2.169E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.073E-06 | 1.971E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.861E-08 | 6.787E-05 |
|---|
| skin development | GO:0043588 |  | 2.25E-07 | 4.104E-04 |
|---|
| replication fork | GO:0005657 |  | 1.203E-06 | 1.463E-03 |
|---|
| response to UV | GO:0009411 |  | 4.638E-06 | 4.23E-03 |
|---|
| enucleate erythrocyte differentiation | GO:0043353 |  | 5.187E-06 | 3.785E-03 |
|---|
| platelet-derived growth factor binding | GO:0048407 |  | 5.187E-06 | 3.154E-03 |
|---|
| ribonuclease H activity | GO:0004523 |  | 5.187E-06 | 2.703E-03 |
|---|
| platelet dense granule | GO:0042827 |  | 5.187E-06 | 2.365E-03 |
|---|
| endodeoxyribonuclease activity. producing 5'-phosphomonoesters | GO:0016888 |  | 5.187E-06 | 2.103E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.187E-06 | 1.892E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 5.187E-06 | 1.72E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 4.853E-07 | 1.168E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.605E-06 | 7.946E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 6.605E-06 | 5.297E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 6.605E-06 | 3.973E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 6.605E-06 | 3.178E-03 |
|---|
| metallo-sulfur cluster assembly | GO:0031163 |  | 6.605E-06 | 2.649E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.526E-06 | 2.93E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 9.903E-06 | 2.978E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 9.903E-06 | 2.647E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 9.903E-06 | 2.383E-03 |
|---|
| response to UV | GO:0009411 |  | 1.152E-05 | 2.519E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 7.794E-07 | 1.793E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 0.01734556 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 0.01156371 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.508E-05 | 8.673E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 2.261E-05 | 0.0103994 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 9.092E-03 |
|---|
| response to UV | GO:0009411 |  | 2.535E-05 | 8.33E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 2.884E-05 | 8.291E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 8.082E-03 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 3.163E-05 | 7.274E-03 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 3.163E-05 | 6.613E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 7.794E-07 | 1.793E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 0.01734556 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 0.01156371 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.508E-05 | 8.673E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 2.261E-05 | 0.0103994 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 9.092E-03 |
|---|
| response to UV | GO:0009411 |  | 2.535E-05 | 8.33E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 2.884E-05 | 8.291E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 8.082E-03 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 3.163E-05 | 7.274E-03 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 3.163E-05 | 6.613E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of DNA replication | GO:0006275 |  | 9.732E-06 | 0.01793563 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.897E-05 | 0.0174829 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.844E-05 | 0.01746934 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.844E-05 | 0.01310201 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 2.844E-05 | 0.0104816 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 2.844E-05 | 8.735E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 3.039E-05 | 8.002E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 3.978E-05 | 9.164E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.978E-05 | 8.146E-03 |
|---|
| negative regulation of lipid metabolic process | GO:0045833 |  | 3.978E-05 | 7.331E-03 |
|---|
| UV protection | GO:0009650 |  | 3.978E-05 | 6.665E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 7.794E-07 | 1.793E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 0.01734556 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 0.01156371 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.508E-05 | 8.673E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 2.261E-05 | 0.0103994 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 9.092E-03 |
|---|
| response to UV | GO:0009411 |  | 2.535E-05 | 8.33E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 2.884E-05 | 8.291E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 8.082E-03 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 3.163E-05 | 7.274E-03 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 3.163E-05 | 6.613E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 1.752E-08 | 4.157E-05 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 6.121E-08 | 7.262E-05 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 1.467E-07 | 1.16E-04 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 1.467E-07 | 8.7E-05 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 2.093E-07 | 9.935E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.093E-07 | 8.279E-05 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 3.831E-07 | 1.299E-04 |
|---|
| negative regulation of lipid metabolic process | GO:0045833 |  | 4.977E-07 | 1.476E-04 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 4.977E-07 | 1.312E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 5.046E-07 | 1.197E-04 |
|---|
| skin development | GO:0043588 |  | 7.904E-07 | 1.705E-04 |
|---|



