Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 890-10-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1671 | 3.453e-02 | 3.034e-02 | 2.231e-01 |
|---|
| IPC-NIBC-129 | 0.2636 | 1.163e-01 | 1.600e-01 | 6.998e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.1892 | 1.292e-02 | 2.102e-02 | 3.128e-01 |
|---|
| Loi_GPL570 | 0.1963 | 1.234e-01 | 1.368e-01 | 4.274e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1690 | 1.000e-06 | 1.300e-05 | 1.000e-06 |
|---|
| Parker_GPL1390 | 0.2280 | 3.541e-02 | 4.470e-02 | 3.244e-01 |
|---|
| Parker_GPL887 | 0.0910 | 4.388e-01 | 4.579e-01 | 4.893e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3289 | 5.849e-01 | 5.165e-01 | 9.835e-01 |
|---|
| Schmidt | 0.2670 | 7.775e-03 | 8.386e-03 | 1.187e-01 |
|---|
| Sotiriou | 0.2884 | 1.801e-02 | 1.402e-02 | 2.615e-01 |
|---|
| Van-De-Vijver | 0.1943 | 6.050e-04 | 8.700e-04 | 4.173e-02 |
|---|
| Zhang | 0.3278 | 1.173e-01 | 7.430e-02 | 2.510e-01 |
|---|
| Zhou | 0.4484 | 8.819e-02 | 7.905e-02 | 4.780e-01 |
|---|
Expression data for subnetwork 890-10-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
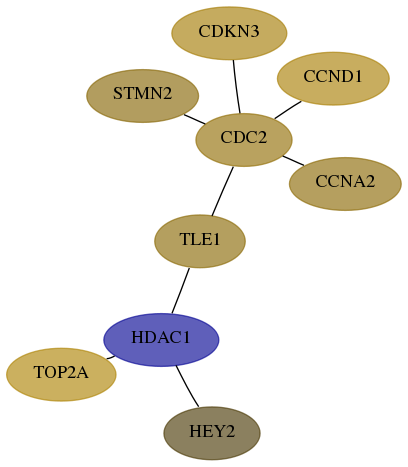
Score for each gene in subnetwork 890-10-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| hdac1 |   | 2 | 78 | 232 | 224 | -0.076 | 0.003 | -0.041 | 0.108 | 0.111 | 0.128 | 0.164 | -0.034 | -0.074 | 0.152 | 0.018 | 0.086 | 0.292 |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| hey2 |   | 1 | 116 | 263 | 263 | 0.013 | 0.189 | -0.017 | 0.127 | 0.076 | 0.165 | -0.287 | -0.039 | 0.152 | 0.076 | -0.071 | -0.043 | 0.367 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| top2a |   | 15 | 11 | 163 | 134 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.147 | 0.234 | 0.273 |
|---|
| ccna2 |   | 1 | 116 | 263 | 263 | 0.065 | 0.183 | 0.157 | 0.177 | 0.133 | 0.079 | 0.026 | 0.276 | 0.264 | 0.176 | 0.170 | 0.173 | 0.404 |
|---|
GO Enrichment output for subnetwork 890-10-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.885E-06 | 0.0204359 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.885E-06 | 0.01021795 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 7.055E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.332E-05 | 7.659E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.545E-05 | 7.108E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 7.144E-03 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 2.484E-05 | 8.16E-03 |
|---|
| DNA ligation | GO:0006266 |  | 3.191E-05 | 9.175E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 3.987E-05 | 0.01018876 |
|---|
| histone deacetylation | GO:0016575 |  | 4.87E-05 | 0.01120123 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.436E-05 | 0.01554808 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.654E-06 | 6.484E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 3.321E-06 | 4.057E-03 |
|---|
| G2/M transition checkpoint | GO:0031576 |  | 3.321E-06 | 2.704E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 4.98E-06 | 3.041E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.095E-06 | 2.489E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.289E-06 | 3.782E-03 |
|---|
| DNA topological change | GO:0006265 |  | 9.289E-06 | 3.242E-03 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 1.194E-05 | 3.646E-03 |
|---|
| DNA ligation | GO:0006266 |  | 1.492E-05 | 4.049E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.492E-05 | 3.644E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.607E-05 | 5.791E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.733E-06 | 8.981E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.185E-06 | 5.034E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 4.185E-06 | 3.356E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.162E-06 | 4.308E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.17E-05 | 5.631E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.17E-05 | 4.693E-03 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 1.504E-05 | 5.17E-03 |
|---|
| DNA ligation | GO:0006266 |  | 1.879E-05 | 5.652E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.879E-05 | 5.024E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.334E-05 | 8.021E-03 |
|---|
| interphase | GO:0051325 |  | 3.659E-05 | 8.002E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.654E-06 | 6.484E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 3.321E-06 | 4.057E-03 |
|---|
| G2/M transition checkpoint | GO:0031576 |  | 3.321E-06 | 2.704E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 4.98E-06 | 3.041E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.095E-06 | 2.489E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.289E-06 | 3.782E-03 |
|---|
| DNA topological change | GO:0006265 |  | 9.289E-06 | 3.242E-03 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 1.194E-05 | 3.646E-03 |
|---|
| DNA ligation | GO:0006266 |  | 1.492E-05 | 4.049E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.492E-05 | 3.644E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.607E-05 | 5.791E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.733E-06 | 8.981E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.185E-06 | 5.034E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 4.185E-06 | 3.356E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.162E-06 | 4.308E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.17E-05 | 5.631E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.17E-05 | 4.693E-03 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 1.504E-05 | 5.17E-03 |
|---|
| DNA ligation | GO:0006266 |  | 1.879E-05 | 5.652E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.879E-05 | 5.024E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.334E-05 | 8.021E-03 |
|---|
| interphase | GO:0051325 |  | 3.659E-05 | 8.002E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.578E-06 | 5.633E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.035E-06 | 3.633E-03 |
|---|
| female pronucleus | GO:0001939 |  | 2.578E-06 | 3.068E-03 |
|---|
| DNA topoisomerase activity | GO:0003916 |  | 2.578E-06 | 2.301E-03 |
|---|
| G2/M transition checkpoint | GO:0031576 |  | 2.578E-06 | 1.841E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 2.578E-06 | 1.534E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.865E-06 | 1.972E-03 |
|---|
| DNA topological change | GO:0006265 |  | 3.865E-06 | 1.725E-03 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 5.41E-06 | 2.147E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 5.41E-06 | 1.932E-03 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 7.212E-06 | 2.341E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 9.742E-06 | 0.03553885 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 9.742E-06 | 0.01776942 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.066E-05 | 0.01296492 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.46E-05 | 0.013319 |
|---|
| DNA topoisomerase activity | GO:0003916 |  | 1.46E-05 | 0.0106552 |
|---|
| G2/M transition checkpoint | GO:0031576 |  | 1.46E-05 | 8.879E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.79E-05 | 9.328E-03 |
|---|
| pronucleus | GO:0045120 |  | 2.043E-05 | 9.318E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.043E-05 | 8.282E-03 |
|---|
| DNA ligation | GO:0006266 |  | 2.043E-05 | 7.454E-03 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.723E-05 | 9.03E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.733E-06 | 8.981E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.185E-06 | 5.034E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 4.185E-06 | 3.356E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.162E-06 | 4.308E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.17E-05 | 5.631E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.17E-05 | 4.693E-03 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 1.504E-05 | 5.17E-03 |
|---|
| DNA ligation | GO:0006266 |  | 1.879E-05 | 5.652E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.879E-05 | 5.024E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.334E-05 | 8.021E-03 |
|---|
| interphase | GO:0051325 |  | 3.659E-05 | 8.002E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.885E-06 | 0.0204359 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.885E-06 | 0.01021795 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 7.055E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.332E-05 | 7.659E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.545E-05 | 7.108E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 7.144E-03 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 2.484E-05 | 8.16E-03 |
|---|
| DNA ligation | GO:0006266 |  | 3.191E-05 | 9.175E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 3.987E-05 | 0.01018876 |
|---|
| histone deacetylation | GO:0016575 |  | 4.87E-05 | 0.01120123 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.436E-05 | 0.01554808 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.885E-06 | 0.0204359 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.885E-06 | 0.01021795 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 7.055E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.332E-05 | 7.659E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.545E-05 | 7.108E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 7.144E-03 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 2.484E-05 | 8.16E-03 |
|---|
| DNA ligation | GO:0006266 |  | 3.191E-05 | 9.175E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 3.987E-05 | 0.01018876 |
|---|
| histone deacetylation | GO:0016575 |  | 4.87E-05 | 0.01120123 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.436E-05 | 0.01554808 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 8.861E-06 | 0.01633162 |
|---|
| negative regulation of signal transduction | GO:0009968 |  | 3.579E-05 | 0.03298143 |
|---|
| histone deacetylation | GO:0016575 |  | 3.978E-05 | 0.02443811 |
|---|
| negative regulation of cell communication | GO:0010648 |  | 4.393E-05 | 0.02023892 |
|---|
| regulation of viral genome replication | GO:0045069 |  | 4.86E-05 | 0.01791259 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.86E-05 | 0.01492716 |
|---|
| vasculogenesis | GO:0001570 |  | 6.885E-05 | 0.01812763 |
|---|
| regulation of viral reproduction | GO:0050792 |  | 6.885E-05 | 0.01586168 |
|---|
| negative regulation of Wnt receptor signaling pathway | GO:0030178 |  | 6.885E-05 | 0.01409927 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 8.029E-05 | 0.01479705 |
|---|
| chromosome condensation | GO:0030261 |  | 9.259E-05 | 0.01551386 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.885E-06 | 0.0204359 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.885E-06 | 0.01021795 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 7.055E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.332E-05 | 7.659E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.545E-05 | 7.108E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 7.144E-03 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 2.484E-05 | 8.16E-03 |
|---|
| DNA ligation | GO:0006266 |  | 3.191E-05 | 9.175E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 3.987E-05 | 0.01018876 |
|---|
| histone deacetylation | GO:0016575 |  | 4.87E-05 | 0.01120123 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.436E-05 | 0.01554808 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 7.724E-06 | 0.01832989 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 7.724E-06 | 9.165E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.055E-06 | 6.372E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.418E-05 | 8.41E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.62E-05 | 7.69E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.62E-05 | 6.409E-03 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 2.159E-05 | 7.32E-03 |
|---|
| DNA ligation | GO:0006266 |  | 3.467E-05 | 0.01028294 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 3.467E-05 | 9.14E-03 |
|---|
| histone deacetylation | GO:0016575 |  | 5.079E-05 | 0.01205238 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.292E-05 | 0.01357304 |
|---|



