Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 7534-9-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1867 | 1.687e-02 | 1.355e-02 | 1.312e-01 |
|---|
| IPC-NIBC-129 | 0.2451 | 5.697e-02 | 8.150e-02 | 4.821e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2244 | 7.663e-03 | 1.305e-02 | 2.348e-01 |
|---|
| Loi_GPL570 | 0.2154 | 9.498e-02 | 1.061e-01 | 3.557e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1696 | 4.000e-06 | 4.100e-05 | 1.400e-05 |
|---|
| Parker_GPL1390 | 0.2521 | 5.806e-02 | 7.162e-02 | 4.270e-01 |
|---|
| Parker_GPL887 | 0.0951 | 3.899e-01 | 4.089e-01 | 4.294e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3578 | 5.648e-01 | 4.950e-01 | 9.803e-01 |
|---|
| Schmidt | 0.2325 | 2.348e-02 | 2.745e-02 | 2.604e-01 |
|---|
| Sotiriou | 0.3165 | 1.129e-02 | 8.668e-03 | 2.019e-01 |
|---|
| Van-De-Vijver | 0.2474 | 1.787e-03 | 2.569e-03 | 8.601e-02 |
|---|
| Zhang | 0.2609 | 1.162e-01 | 7.344e-02 | 2.491e-01 |
|---|
| Zhou | 0.3878 | 6.781e-02 | 5.630e-02 | 3.973e-01 |
|---|
Expression data for subnetwork 7534-9-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
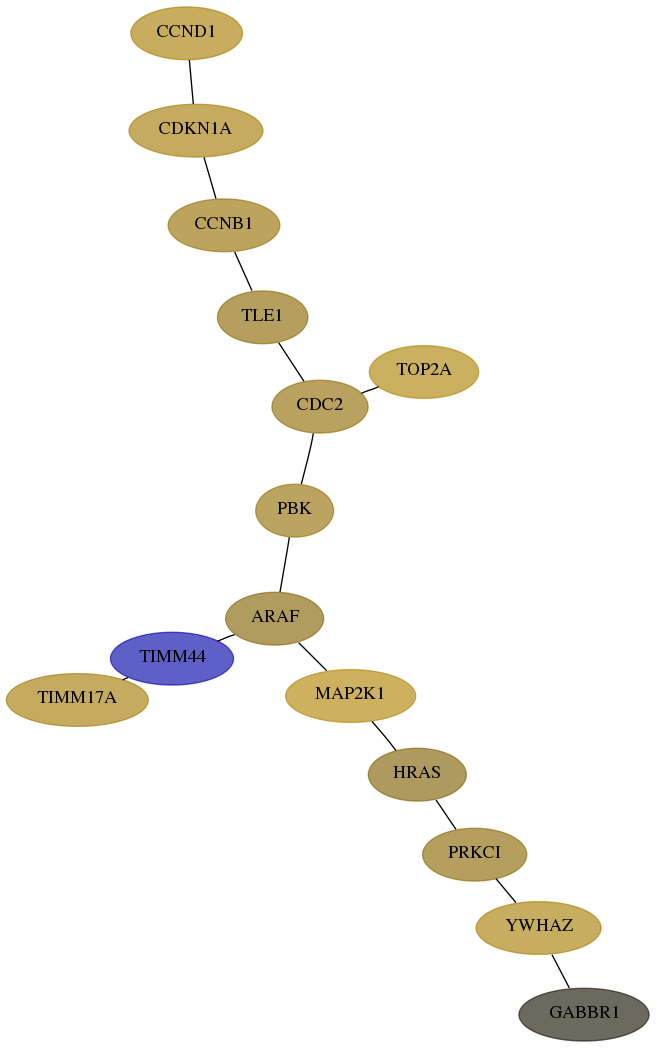
Score for each gene in subnetwork 7534-9-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| araf |   | 1 | 116 | 242 | 244 | 0.055 | -0.065 | 0.131 | 0.017 | 0.080 | 0.133 | 0.377 | -0.081 | 0.030 | -0.050 | 0.097 | -0.124 | 0.167 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| ywhaz |   | 17 | 10 | 29 | 25 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | undef | 0.103 | 0.213 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| prkci |   | 14 | 15 | 31 | 26 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | 0.049 | 0.124 | 0.149 |
|---|
| timm17a |   | 1 | 116 | 242 | 244 | 0.112 | 0.301 | 0.144 | 0.162 | 0.173 | 0.140 | 0.210 | 0.358 | 0.147 | 0.160 | 0.068 | 0.113 | 0.255 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| pbk |   | 1 | 116 | 242 | 244 | 0.081 | 0.245 | 0.138 | 0.065 | 0.167 | 0.139 | 0.135 | 0.214 | 0.183 | 0.193 | 0.122 | 0.169 | 0.303 |
|---|
| timm44 |   | 1 | 116 | 242 | 244 | -0.127 | 0.211 | 0.022 | 0.244 | 0.072 | 0.234 | -0.080 | 0.180 | -0.052 | -0.079 | 0.128 | 0.136 | -0.010 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| hras |   | 14 | 15 | 42 | 34 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.195 | 0.022 | 0.245 | 0.086 | 0.208 | 0.180 | 0.003 | 0.197 |
|---|
| gabbr1 |   | 7 | 30 | 56 | 49 | 0.003 | -0.016 | -0.022 | -0.051 | 0.082 | 0.119 | 0.584 | -0.082 | 0.049 | 0.057 | 0.125 | 0.096 | 0.116 |
|---|
| top2a |   | 15 | 11 | 163 | 134 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.147 | 0.234 | 0.273 |
|---|
| map2k1 |   | 10 | 20 | 54 | 44 | 0.154 | 0.096 | 0.093 | 0.013 | 0.095 | 0.045 | -0.223 | 0.207 | 0.082 | 0.096 | undef | undef | 0.230 |
|---|
GO Enrichment output for subnetwork 7534-9-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 1.046E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 8.573E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 1.929E-03 |
|---|
| protein localization in mitochondrion | GO:0070585 |  | 2.515E-06 | 1.446E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.097E-06 | 2.345E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.94E-06 | 2.277E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.008E-06 | 2.96E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.357E-05 | 3.901E-03 |
|---|
| interphase | GO:0051325 |  | 1.486E-05 | 3.798E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.617E-05 | 3.719E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 2.097E-05 | 4.385E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 9.528E-09 | 2.328E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.71E-07 | 3.31E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 5.295E-07 | 4.312E-04 |
|---|
| protein localization in mitochondrion | GO:0070585 |  | 1.075E-06 | 6.565E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.253E-06 | 6.123E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.161E-06 | 8.799E-04 |
|---|
| osteoblast differentiation | GO:0001649 |  | 2.441E-06 | 8.52E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.441E-06 | 7.455E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.991E-06 | 8.118E-04 |
|---|
| sperm motility | GO:0030317 |  | 3.072E-06 | 7.504E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 4.638E-06 | 1.03E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.566E-08 | 3.767E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.449E-07 | 5.352E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 8.69E-07 | 6.969E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.763E-06 | 1.061E-03 |
|---|
| protein localization in mitochondrion | GO:0070585 |  | 1.763E-06 | 8.484E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.12E-06 | 1.251E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.543E-06 | 1.218E-03 |
|---|
| sperm motility | GO:0030317 |  | 3.543E-06 | 1.065E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 3.543E-06 | 9.471E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.064E-06 | 1.218E-03 |
|---|
| interphase | GO:0051325 |  | 5.732E-06 | 1.254E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 9.528E-09 | 2.328E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.71E-07 | 3.31E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 5.295E-07 | 4.312E-04 |
|---|
| protein localization in mitochondrion | GO:0070585 |  | 1.075E-06 | 6.565E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.253E-06 | 6.123E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.161E-06 | 8.799E-04 |
|---|
| osteoblast differentiation | GO:0001649 |  | 2.441E-06 | 8.52E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.441E-06 | 7.455E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.991E-06 | 8.118E-04 |
|---|
| sperm motility | GO:0030317 |  | 3.072E-06 | 7.504E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 4.638E-06 | 1.03E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.566E-08 | 3.767E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.449E-07 | 5.352E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 8.69E-07 | 6.969E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.763E-06 | 1.061E-03 |
|---|
| protein localization in mitochondrion | GO:0070585 |  | 1.763E-06 | 8.484E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.12E-06 | 1.251E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.543E-06 | 1.218E-03 |
|---|
| sperm motility | GO:0030317 |  | 3.543E-06 | 1.065E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 3.543E-06 | 9.471E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.064E-06 | 1.218E-03 |
|---|
| interphase | GO:0051325 |  | 5.732E-06 | 1.254E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 3.814E-08 | 1.362E-04 |
|---|
| macromolecule transmembrane transporter activity | GO:0022884 |  | 8.163E-08 | 1.458E-04 |
|---|
| mitochondrial inner membrane presequence translocase complex | GO:0005744 |  | 8.163E-08 | 9.717E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.122E-07 | 1.001E-04 |
|---|
| protein transmembrane transporter activity | GO:0008320 |  | 3.087E-07 | 2.204E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.608E-07 | 2.742E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.549E-07 | 4.361E-04 |
|---|
| protein localization in mitochondrion | GO:0070585 |  | 1.552E-06 | 6.926E-04 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.371E-06 | 3.321E-03 |
|---|
| DNA topoisomerase activity | GO:0003916 |  | 8.371E-06 | 2.989E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.012E-05 | 3.285E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.325E-08 | 8.482E-05 |
|---|
| macromolecule transmembrane transporter activity | GO:0022884 |  | 4.978E-08 | 9.079E-05 |
|---|
| mitochondrial inner membrane presequence translocase complex | GO:0005744 |  | 4.978E-08 | 6.053E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.507E-07 | 1.374E-04 |
|---|
| protein transmembrane transporter activity | GO:0008320 |  | 1.883E-07 | 1.374E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.316E-07 | 1.408E-04 |
|---|
| protein localization in mitochondrion | GO:0070585 |  | 2.811E-07 | 1.465E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.282E-07 | 1.952E-04 |
|---|
| interphase | GO:0051325 |  | 4.725E-07 | 1.915E-04 |
|---|
| response to light stimulus | GO:0009416 |  | 1E-06 | 3.648E-04 |
|---|
| mitochondrial transport | GO:0006839 |  | 4.689E-06 | 1.555E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.566E-08 | 3.767E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.449E-07 | 5.352E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 8.69E-07 | 6.969E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.763E-06 | 1.061E-03 |
|---|
| protein localization in mitochondrion | GO:0070585 |  | 1.763E-06 | 8.484E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.12E-06 | 1.251E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.543E-06 | 1.218E-03 |
|---|
| sperm motility | GO:0030317 |  | 3.543E-06 | 1.065E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 3.543E-06 | 9.471E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.064E-06 | 1.218E-03 |
|---|
| interphase | GO:0051325 |  | 5.732E-06 | 1.254E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 1.046E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 8.573E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 1.929E-03 |
|---|
| protein localization in mitochondrion | GO:0070585 |  | 2.515E-06 | 1.446E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.097E-06 | 2.345E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.94E-06 | 2.277E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.008E-06 | 2.96E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.357E-05 | 3.901E-03 |
|---|
| interphase | GO:0051325 |  | 1.486E-05 | 3.798E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.617E-05 | 3.719E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 2.097E-05 | 4.385E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 1.046E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 8.573E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 1.929E-03 |
|---|
| protein localization in mitochondrion | GO:0070585 |  | 2.515E-06 | 1.446E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.097E-06 | 2.345E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.94E-06 | 2.277E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.008E-06 | 2.96E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.357E-05 | 3.901E-03 |
|---|
| interphase | GO:0051325 |  | 1.486E-05 | 3.798E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.617E-05 | 3.719E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 2.097E-05 | 4.385E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein targeting to mitochondrion | GO:0006626 |  | 2.64E-07 | 4.866E-04 |
|---|
| mitochondrial transport | GO:0006839 |  | 4.406E-06 | 4.06E-03 |
|---|
| regulation of long-term neuronal synaptic plasticity | GO:0048169 |  | 1.897E-05 | 0.01165527 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 1.897E-05 | 8.741E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.897E-05 | 6.993E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 1.897E-05 | 5.828E-03 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.897E-05 | 4.995E-03 |
|---|
| intracellular protein transmembrane transport | GO:0065002 |  | 1.967E-05 | 4.53E-03 |
|---|
| mitochondrion organization | GO:0007005 |  | 2.647E-05 | 5.421E-03 |
|---|
| establishment or maintenance of apical/basal cell polarity | GO:0035088 |  | 2.844E-05 | 5.241E-03 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 2.844E-05 | 4.764E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 1.046E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 8.573E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 1.929E-03 |
|---|
| protein localization in mitochondrion | GO:0070585 |  | 2.515E-06 | 1.446E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.097E-06 | 2.345E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.94E-06 | 2.277E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.008E-06 | 2.96E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.357E-05 | 3.901E-03 |
|---|
| interphase | GO:0051325 |  | 1.486E-05 | 3.798E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.617E-05 | 3.719E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 2.097E-05 | 4.385E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 3.422E-08 | 8.12E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.473E-07 | 8.866E-04 |
|---|
| protein localization in mitochondrion | GO:0070585 |  | 1.894E-06 | 1.498E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.298E-06 | 1.363E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.475E-06 | 2.124E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.176E-06 | 2.047E-03 |
|---|
| sperm motility | GO:0030317 |  | 8.702E-06 | 2.95E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.821E-06 | 2.913E-03 |
|---|
| interphase | GO:0051325 |  | 1.075E-05 | 2.834E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.496E-05 | 3.551E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.986E-05 | 4.284E-03 |
|---|



