Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6426-8-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1549 | 1.423e-02 | 1.117e-02 | 1.151e-01 |
|---|
| IPC-NIBC-129 | 0.2651 | 6.951e-02 | 9.847e-02 | 5.411e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2327 | 7.210e-03 | 1.234e-02 | 2.268e-01 |
|---|
| Loi_GPL570 | 0.2344 | 7.295e-02 | 8.215e-02 | 2.928e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1745 | 1.000e-06 | 1.200e-05 | 1.000e-06 |
|---|
| Parker_GPL1390 | 0.2549 | 4.363e-02 | 5.455e-02 | 3.653e-01 |
|---|
| Parker_GPL887 | 0.0936 | 4.717e-01 | 4.907e-01 | 5.291e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3578 | 5.718e-01 | 5.025e-01 | 9.814e-01 |
|---|
| Schmidt | 0.2525 | 1.770e-02 | 2.031e-02 | 2.152e-01 |
|---|
| Sotiriou | 0.2873 | 1.834e-02 | 1.429e-02 | 2.641e-01 |
|---|
| Van-De-Vijver | 0.2327 | 1.083e-03 | 1.558e-03 | 6.190e-02 |
|---|
| Zhang | 0.2781 | 1.078e-01 | 6.682e-02 | 2.339e-01 |
|---|
| Zhou | 0.4159 | 6.781e-02 | 5.630e-02 | 3.973e-01 |
|---|
Expression data for subnetwork 6426-8-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
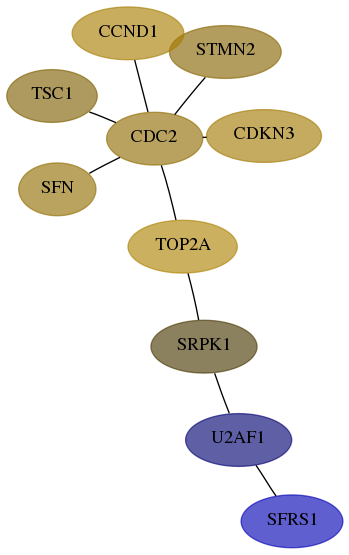
Score for each gene in subnetwork 6426-8-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| sfrs1 |   | 1 | 116 | 224 | 227 | -0.174 | 0.093 | 0.077 | -0.042 | 0.111 | 0.166 | -0.306 | 0.206 | 0.084 | 0.138 | undef | -0.029 | 0.245 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| srpk1 |   | 3 | 62 | 178 | 167 | 0.013 | 0.261 | 0.105 | 0.234 | 0.112 | 0.272 | -0.071 | 0.144 | 0.241 | 0.115 | 0.035 | 0.050 | 0.193 |
|---|
| u2af1 |   | 2 | 78 | 178 | 173 | -0.037 | 0.160 | 0.147 | 0.045 | 0.111 | 0.190 | 0.120 | 0.214 | 0.257 | 0.007 | 0.132 | -0.070 | 0.159 |
|---|
| top2a |   | 15 | 11 | 163 | 134 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.147 | 0.234 | 0.273 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
GO Enrichment output for subnetwork 6426-8-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 3.419E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.885E-06 | 0.01021795 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.885E-06 | 6.812E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.885E-06 | 5.109E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 8.885E-06 | 4.087E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 3.527E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.332E-05 | 4.377E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 5.358E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.864E-05 | 4.763E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 2.163E-05 | 4.976E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.484E-05 | 5.193E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.329E-08 | 1.79E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.573E-06 | 5.586E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.743E-06 | 3.863E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.743E-06 | 2.897E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 7.112E-06 | 3.475E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.112E-06 | 2.896E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.326E-05 | 4.629E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.326E-05 | 4.05E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.326E-05 | 3.6E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 1.429E-05 | 3.492E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.705E-05 | 3.786E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.191E-07 | 2.865E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.077E-06 | 7.311E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.077E-06 | 4.874E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.077E-06 | 3.655E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 6.077E-06 | 2.924E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.599E-06 | 2.646E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.699E-05 | 5.84E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.699E-05 | 5.11E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.699E-05 | 4.542E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 2.06E-05 | 4.956E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.183E-05 | 4.776E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.329E-08 | 1.79E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.573E-06 | 5.586E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.743E-06 | 3.863E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.743E-06 | 2.897E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 7.112E-06 | 3.475E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.112E-06 | 2.896E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.326E-05 | 4.629E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.326E-05 | 4.05E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.326E-05 | 3.6E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 1.429E-05 | 3.492E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.705E-05 | 3.786E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.191E-07 | 2.865E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.077E-06 | 7.311E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.077E-06 | 4.874E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.077E-06 | 3.655E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 6.077E-06 | 2.924E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.599E-06 | 2.646E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.699E-05 | 5.84E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.699E-05 | 5.11E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.699E-05 | 4.542E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 2.06E-05 | 4.956E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.183E-05 | 4.776E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.46E-08 | 5.213E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.361E-06 | 4.216E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 3.314E-06 | 3.944E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.314E-06 | 2.958E-03 |
|---|
| DNA topoisomerase activity | GO:0003916 |  | 3.314E-06 | 2.367E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 4.536E-06 | 2.7E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.969E-06 | 2.535E-03 |
|---|
| DNA topological change | GO:0006265 |  | 4.969E-06 | 2.218E-03 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 6.445E-06 | 2.557E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 6.954E-06 | 2.483E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.954E-06 | 2.258E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.59E-08 | 5.799E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.839E-06 | 3.354E-03 |
|---|
| RS domain binding | GO:0050733 |  | 3.136E-06 | 3.813E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 3.136E-06 | 2.86E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 3.136E-06 | 2.288E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 3.136E-06 | 1.907E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 4.34E-06 | 2.262E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.703E-06 | 2.144E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.703E-06 | 1.906E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.703E-06 | 1.716E-03 |
|---|
| DNA topoisomerase activity | GO:0003916 |  | 4.703E-06 | 1.56E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.191E-07 | 2.865E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.077E-06 | 7.311E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.077E-06 | 4.874E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.077E-06 | 3.655E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 6.077E-06 | 2.924E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.599E-06 | 2.646E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.699E-05 | 5.84E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.699E-05 | 5.11E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.699E-05 | 4.542E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 2.06E-05 | 4.956E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.183E-05 | 4.776E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 3.419E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.885E-06 | 0.01021795 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.885E-06 | 6.812E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.885E-06 | 5.109E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 8.885E-06 | 4.087E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 3.527E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.332E-05 | 4.377E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 5.358E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.864E-05 | 4.763E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 2.163E-05 | 4.976E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.484E-05 | 5.193E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 3.419E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.885E-06 | 0.01021795 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.885E-06 | 6.812E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.885E-06 | 5.109E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 8.885E-06 | 4.087E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 3.527E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.332E-05 | 4.377E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 5.358E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.864E-05 | 4.763E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 2.163E-05 | 4.976E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.484E-05 | 5.193E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromosome segregation | GO:0007059 |  | 5.36E-06 | 9.878E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 8.861E-06 | 8.166E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.329E-05 | 8.162E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.329E-05 | 6.121E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 1.329E-05 | 4.897E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 1.329E-05 | 4.081E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 1.859E-05 | 4.895E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.859E-05 | 4.283E-03 |
|---|
| regulation of mRNA processing | GO:0050684 |  | 1.859E-05 | 3.807E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 2.478E-05 | 4.566E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.478E-05 | 4.151E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 3.419E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.885E-06 | 0.01021795 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 8.885E-06 | 6.812E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.885E-06 | 5.109E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 8.885E-06 | 4.087E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 3.527E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.332E-05 | 4.377E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 5.358E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.864E-05 | 4.763E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 2.163E-05 | 4.976E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.484E-05 | 5.193E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.601E-07 | 6.171E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.06E-05 | 0.01257554 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.06E-05 | 8.384E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.06E-05 | 6.288E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.307E-05 | 6.202E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.589E-05 | 6.284E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.223E-05 | 7.536E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.223E-05 | 6.594E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.962E-05 | 7.81E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.962E-05 | 7.029E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 3.175E-05 | 6.85E-03 |
|---|



