Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 64130-8-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1724 | 1.177e-02 | 9.009e-03 | 9.918e-02 |
|---|
| IPC-NIBC-129 | 0.2458 | 2.779e-02 | 4.096e-02 | 2.983e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2419 | 5.158e-03 | 9.085e-03 | 1.865e-01 |
|---|
| Loi_GPL570 | 0.2629 | 4.879e-02 | 5.555e-02 | 2.143e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1604 | 4.000e-06 | 3.900e-05 | 1.300e-05 |
|---|
| Parker_GPL1390 | 0.2702 | 2.909e-02 | 3.705e-02 | 2.890e-01 |
|---|
| Parker_GPL887 | 0.1917 | 4.251e-01 | 4.443e-01 | 4.726e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3421 | 2.241e-01 | 1.628e-01 | 7.865e-01 |
|---|
| Schmidt | 0.2805 | 6.589e-02 | 8.145e-02 | 4.854e-01 |
|---|
| Sotiriou | 0.3893 | 3.320e-03 | 2.463e-03 | 9.770e-02 |
|---|
| Van-De-Vijver | 0.2996 | 1.144e-03 | 1.646e-03 | 6.421e-02 |
|---|
| Zhang | 0.1973 | 1.335e-01 | 8.749e-02 | 2.796e-01 |
|---|
| Zhou | 0.4128 | 7.827e-02 | 6.781e-02 | 4.404e-01 |
|---|
Expression data for subnetwork 64130-8-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
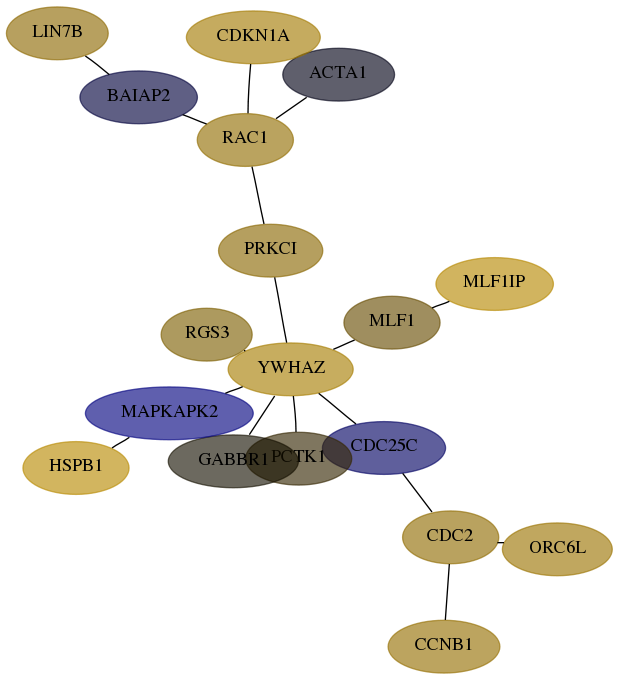
Score for each gene in subnetwork 64130-8-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| rgs3 |   | 4 | 50 | 77 | 66 | 0.048 | 0.195 | 0.029 | 0.052 | 0.056 | 0.192 | -0.052 | -0.078 | 0.033 | 0.023 | undef | 0.100 | -0.196 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| lin7b |   | 1 | 116 | 98 | 105 | 0.067 | 0.057 | 0.096 | 0.131 | 0.094 | -0.102 | 0.082 | 0.070 | 0.163 | 0.053 | undef | 0.149 | 0.069 |
|---|
| mlf1ip |   | 5 | 40 | 77 | 64 | 0.182 | 0.147 | 0.186 | 0.144 | 0.242 | 0.266 | 0.005 | 0.345 | 0.182 | 0.197 | undef | 0.240 | 0.372 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| prkci |   | 14 | 15 | 31 | 26 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | 0.049 | 0.124 | 0.149 |
|---|
| orc6l |   | 15 | 11 | 34 | 28 | 0.094 | 0.270 | 0.084 | 0.114 | 0.106 | 0.240 | 0.256 | 0.234 | 0.192 | 0.105 | 0.258 | 0.175 | 0.160 |
|---|
| rac1 |   | 1 | 116 | 98 | 105 | 0.076 | 0.125 | 0.034 | -0.009 | 0.093 | -0.271 | -0.507 | 0.060 | -0.044 | 0.062 | 0.116 | -0.141 | 0.202 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| gabbr1 |   | 7 | 30 | 56 | 49 | 0.003 | -0.016 | -0.022 | -0.051 | 0.082 | 0.119 | 0.584 | -0.082 | 0.049 | 0.057 | 0.125 | 0.096 | 0.116 |
|---|
| baiap2 |   | 1 | 116 | 98 | 105 | -0.009 | -0.090 | 0.049 | 0.050 | 0.114 | 0.021 | -0.194 | 0.101 | 0.130 | 0.140 | 0.065 | 0.167 | 0.049 |
|---|
| mlf1 |   | 7 | 30 | 56 | 49 | 0.027 | 0.224 | 0.053 | 0.047 | 0.169 | 0.128 | 0.021 | -0.028 | 0.106 | 0.176 | undef | 0.098 | 0.141 |
|---|
| cdc25c |   | 2 | 78 | 81 | 80 | -0.025 | 0.187 | 0.084 | 0.267 | 0.070 | 0.059 | 0.367 | 0.125 | 0.204 | 0.250 | 0.086 | 0.054 | 0.351 |
|---|
| ywhaz |   | 17 | 10 | 29 | 25 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | undef | 0.103 | 0.213 |
|---|
| hspb1 |   | 5 | 40 | 42 | 36 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.183 | -0.077 | 0.290 |
|---|
| mapkapk2 |   | 3 | 62 | 42 | 45 | -0.050 | 0.102 | 0.156 | -0.116 | 0.090 | 0.168 | 0.303 | 0.074 | 0.171 | 0.117 | 0.092 | -0.124 | 0.312 |
|---|
| pctk1 |   | 1 | 116 | 98 | 105 | 0.008 | 0.100 | 0.114 | 0.157 | 0.107 | -0.240 | -0.303 | 0.010 | 0.044 | 0.143 | undef | 0.128 | 0.250 |
|---|
| acta1 |   | 9 | 22 | 1 | 7 | -0.003 | 0.098 | -0.008 | -0.030 | 0.119 | 0.174 | 0.376 | 0.024 | 0.182 | 0.214 | 0.100 | 0.048 | 0.201 |
|---|
GO Enrichment output for subnetwork 64130-8-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 7.997E-08 | 1.839E-04 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 7.997E-08 | 9.196E-05 |
|---|
| actin filament organization | GO:0007015 |  | 1.281E-07 | 9.823E-05 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 1.597E-07 | 9.183E-05 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 4.459E-07 | 2.051E-04 |
|---|
| ruffle organization | GO:0031529 |  | 4.459E-07 | 1.709E-04 |
|---|
| lamellipodium assembly | GO:0030032 |  | 4.459E-07 | 1.465E-04 |
|---|
| localization within membrane | GO:0051668 |  | 6.679E-07 | 1.92E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.308E-06 | 3.343E-04 |
|---|
| actin filament polymerization | GO:0030041 |  | 1.308E-06 | 3.009E-04 |
|---|
| cell projection assembly | GO:0030031 |  | 4.126E-06 | 8.628E-04 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of receptor-mediated endocytosis | GO:0048261 |  | 1.462E-11 | 3.571E-08 |
|---|
| regulation of hydrogen peroxide metabolic process | GO:0010310 |  | 4.381E-11 | 5.351E-08 |
|---|
| regulation of oxygen and reactive oxygen species metabolic process | GO:0080010 |  | 1.021E-10 | 8.316E-08 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 2.04E-10 | 1.246E-07 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 3.669E-10 | 1.793E-07 |
|---|
| ruffle organization | GO:0031529 |  | 6.109E-10 | 2.487E-07 |
|---|
| actin filament organization | GO:0007015 |  | 6.405E-10 | 2.236E-07 |
|---|
| lamellipodium assembly | GO:0030032 |  | 1.437E-09 | 4.388E-07 |
|---|
| localization within membrane | GO:0051668 |  | 1.437E-09 | 3.901E-07 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 2.9E-09 | 7.086E-07 |
|---|
| actin filament polymerization | GO:0030041 |  | 3.951E-09 | 8.775E-07 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 3.111E-08 | 7.484E-05 |
|---|
| regulation of hydrogen peroxide metabolic process | GO:0010310 |  | 3.111E-08 | 3.742E-05 |
|---|
| actin filament organization | GO:0007015 |  | 4.944E-08 | 3.965E-05 |
|---|
| regulation of oxygen and reactive oxygen species metabolic process | GO:0080010 |  | 6.215E-08 | 3.738E-05 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 1.086E-07 | 5.228E-05 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 1.736E-07 | 6.963E-05 |
|---|
| ruffle organization | GO:0031529 |  | 2.602E-07 | 8.943E-05 |
|---|
| localization within membrane | GO:0051668 |  | 3.713E-07 | 1.117E-04 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 5.1E-07 | 1.363E-04 |
|---|
| lamellipodium assembly | GO:0030032 |  | 5.1E-07 | 1.227E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 8.822E-07 | 1.93E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of receptor-mediated endocytosis | GO:0048261 |  | 1.462E-11 | 3.571E-08 |
|---|
| regulation of hydrogen peroxide metabolic process | GO:0010310 |  | 4.381E-11 | 5.351E-08 |
|---|
| regulation of oxygen and reactive oxygen species metabolic process | GO:0080010 |  | 1.021E-10 | 8.316E-08 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 2.04E-10 | 1.246E-07 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 3.669E-10 | 1.793E-07 |
|---|
| ruffle organization | GO:0031529 |  | 6.109E-10 | 2.487E-07 |
|---|
| actin filament organization | GO:0007015 |  | 6.405E-10 | 2.236E-07 |
|---|
| lamellipodium assembly | GO:0030032 |  | 1.437E-09 | 4.388E-07 |
|---|
| localization within membrane | GO:0051668 |  | 1.437E-09 | 3.901E-07 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 2.9E-09 | 7.086E-07 |
|---|
| actin filament polymerization | GO:0030041 |  | 3.951E-09 | 8.775E-07 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 3.111E-08 | 7.484E-05 |
|---|
| regulation of hydrogen peroxide metabolic process | GO:0010310 |  | 3.111E-08 | 3.742E-05 |
|---|
| actin filament organization | GO:0007015 |  | 4.944E-08 | 3.965E-05 |
|---|
| regulation of oxygen and reactive oxygen species metabolic process | GO:0080010 |  | 6.215E-08 | 3.738E-05 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 1.086E-07 | 5.228E-05 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 1.736E-07 | 6.963E-05 |
|---|
| ruffle organization | GO:0031529 |  | 2.602E-07 | 8.943E-05 |
|---|
| localization within membrane | GO:0051668 |  | 3.713E-07 | 1.117E-04 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 5.1E-07 | 1.363E-04 |
|---|
| lamellipodium assembly | GO:0030032 |  | 5.1E-07 | 1.227E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 8.822E-07 | 1.93E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| actin filament organization | GO:0007015 |  | 5.504E-07 | 1.965E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.03E-06 | 1.839E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.569E-06 | 3.059E-03 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 1.407E-05 | 0.01255729 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 1.407E-05 | 0.01004583 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 2.108E-05 | 0.01254819 |
|---|
| nuclear origin of replication recognition complex | GO:0005664 |  | 2.108E-05 | 0.0107556 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 2.108E-05 | 9.411E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 2.108E-05 | 8.365E-03 |
|---|
| actin filament polymerization | GO:0030041 |  | 2.108E-05 | 7.529E-03 |
|---|
| ruffle organization | GO:0031529 |  | 2.108E-05 | 6.844E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| actin filament organization | GO:0007015 |  | 7.534E-07 | 2.748E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 8.978E-07 | 1.638E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.705E-06 | 3.29E-03 |
|---|
| interphase | GO:0051325 |  | 2.983E-06 | 2.721E-03 |
|---|
| cyclin-dependent protein kinase activity | GO:0004693 |  | 4.648E-06 | 3.391E-03 |
|---|
| negative regulation of receptor-mediated endocytosis | GO:0048261 |  | 1.441E-05 | 8.76E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 1.441E-05 | 7.508E-03 |
|---|
| muscle thin filament assembly | GO:0030240 |  | 1.441E-05 | 6.57E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 1.441E-05 | 5.84E-03 |
|---|
| origin recognition complex | GO:0000808 |  | 1.441E-05 | 5.256E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 1.441E-05 | 4.778E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 3.111E-08 | 7.484E-05 |
|---|
| regulation of hydrogen peroxide metabolic process | GO:0010310 |  | 3.111E-08 | 3.742E-05 |
|---|
| actin filament organization | GO:0007015 |  | 4.944E-08 | 3.965E-05 |
|---|
| regulation of oxygen and reactive oxygen species metabolic process | GO:0080010 |  | 6.215E-08 | 3.738E-05 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 1.086E-07 | 5.228E-05 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 1.736E-07 | 6.963E-05 |
|---|
| ruffle organization | GO:0031529 |  | 2.602E-07 | 8.943E-05 |
|---|
| localization within membrane | GO:0051668 |  | 3.713E-07 | 1.117E-04 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 5.1E-07 | 1.363E-04 |
|---|
| lamellipodium assembly | GO:0030032 |  | 5.1E-07 | 1.227E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 8.822E-07 | 1.93E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 7.997E-08 | 1.839E-04 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 7.997E-08 | 9.196E-05 |
|---|
| actin filament organization | GO:0007015 |  | 1.281E-07 | 9.823E-05 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 1.597E-07 | 9.183E-05 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 4.459E-07 | 2.051E-04 |
|---|
| ruffle organization | GO:0031529 |  | 4.459E-07 | 1.709E-04 |
|---|
| lamellipodium assembly | GO:0030032 |  | 4.459E-07 | 1.465E-04 |
|---|
| localization within membrane | GO:0051668 |  | 6.679E-07 | 1.92E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.308E-06 | 3.343E-04 |
|---|
| actin filament polymerization | GO:0030041 |  | 1.308E-06 | 3.009E-04 |
|---|
| cell projection assembly | GO:0030031 |  | 4.126E-06 | 8.628E-04 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 7.997E-08 | 1.839E-04 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 7.997E-08 | 9.196E-05 |
|---|
| actin filament organization | GO:0007015 |  | 1.281E-07 | 9.823E-05 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 1.597E-07 | 9.183E-05 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 4.459E-07 | 2.051E-04 |
|---|
| ruffle organization | GO:0031529 |  | 4.459E-07 | 1.709E-04 |
|---|
| lamellipodium assembly | GO:0030032 |  | 4.459E-07 | 1.465E-04 |
|---|
| localization within membrane | GO:0051668 |  | 6.679E-07 | 1.92E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.308E-06 | 3.343E-04 |
|---|
| actin filament polymerization | GO:0030041 |  | 1.308E-06 | 3.009E-04 |
|---|
| cell projection assembly | GO:0030031 |  | 4.126E-06 | 8.628E-04 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| actin filament organization | GO:0007015 |  | 6.641E-07 | 1.224E-03 |
|---|
| cell projection assembly | GO:0030031 |  | 5.293E-06 | 4.877E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 2.318E-05 | 0.01424118 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 2.318E-05 | 0.01068088 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 2.318E-05 | 8.545E-03 |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.318E-05 | 7.121E-03 |
|---|
| ruffle organization | GO:0031529 |  | 3.474E-05 | 9.147E-03 |
|---|
| establishment or maintenance of apical/basal cell polarity | GO:0035088 |  | 3.474E-05 | 8.004E-03 |
|---|
| localization within membrane | GO:0051668 |  | 3.474E-05 | 7.114E-03 |
|---|
| regulation of receptor-mediated endocytosis | GO:0048259 |  | 4.86E-05 | 8.956E-03 |
|---|
| myofibril assembly | GO:0030239 |  | 4.86E-05 | 8.142E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 7.997E-08 | 1.839E-04 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 7.997E-08 | 9.196E-05 |
|---|
| actin filament organization | GO:0007015 |  | 1.281E-07 | 9.823E-05 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 1.597E-07 | 9.183E-05 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 4.459E-07 | 2.051E-04 |
|---|
| ruffle organization | GO:0031529 |  | 4.459E-07 | 1.709E-04 |
|---|
| lamellipodium assembly | GO:0030032 |  | 4.459E-07 | 1.465E-04 |
|---|
| localization within membrane | GO:0051668 |  | 6.679E-07 | 1.92E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.308E-06 | 3.343E-04 |
|---|
| actin filament polymerization | GO:0030041 |  | 1.308E-06 | 3.009E-04 |
|---|
| cell projection assembly | GO:0030031 |  | 4.126E-06 | 8.628E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.981E-08 | 1.182E-04 |
|---|
| actin filament organization | GO:0007015 |  | 8.868E-08 | 1.052E-04 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 9.95E-08 | 7.87E-05 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 9.95E-08 | 5.903E-05 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 2.779E-07 | 1.319E-04 |
|---|
| ruffle organization | GO:0031529 |  | 2.779E-07 | 1.099E-04 |
|---|
| lamellipodium assembly | GO:0030032 |  | 4.164E-07 | 1.412E-04 |
|---|
| localization within membrane | GO:0051668 |  | 4.164E-07 | 1.235E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.087E-06 | 2.865E-04 |
|---|
| actin filament polymerization | GO:0030041 |  | 1.411E-06 | 3.348E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.338E-06 | 7.202E-04 |
|---|



