Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6227-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1696 | 1.545e-02 | 1.226e-02 | 1.226e-01 |
|---|
| IPC-NIBC-129 | 0.2256 | 6.767e-02 | 9.600e-02 | 5.331e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2287 | 1.819e-02 | 2.872e-02 | 3.731e-01 |
|---|
| Loi_GPL570 | 0.2522 | 5.679e-02 | 6.439e-02 | 2.416e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1708 | 1.600e-05 | 1.320e-04 | 1.340e-04 |
|---|
| Parker_GPL1390 | 0.2121 | 8.623e-02 | 1.043e-01 | 5.225e-01 |
|---|
| Parker_GPL887 | 0.1445 | 4.322e-01 | 4.513e-01 | 4.812e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3370 | 3.593e-01 | 2.867e-01 | 9.092e-01 |
|---|
| Schmidt | 0.2046 | 1.927e-02 | 2.224e-02 | 2.280e-01 |
|---|
| Sotiriou | 0.2659 | 2.608e-02 | 2.055e-02 | 3.182e-01 |
|---|
| Van-De-Vijver | 0.2347 | 9.360e-04 | 1.348e-03 | 5.616e-02 |
|---|
| Zhang | 0.2730 | 1.141e-01 | 7.177e-02 | 2.453e-01 |
|---|
| Zhou | 0.4240 | 8.196e-02 | 7.195e-02 | 4.547e-01 |
|---|
Expression data for subnetwork 6227-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
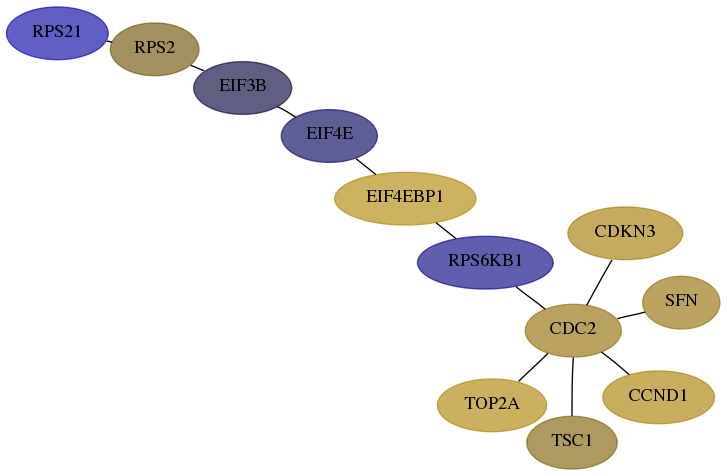
Score for each gene in subnetwork 6227-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| rps6kb1 |   | 5 | 40 | 216 | 200 | -0.048 | 0.004 | 0.033 | 0.144 | 0.128 | 0.095 | -0.319 | 0.022 | 0.116 | 0.084 | 0.068 | 0.076 | 0.280 |
|---|
| eif4ebp1 |   | 4 | 50 | 260 | 239 | 0.158 | 0.203 | 0.132 | 0.139 | 0.104 | 0.122 | 0.119 | 0.252 | 0.031 | 0.042 | 0.188 | 0.029 | 0.219 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| rps2 |   | 1 | 116 | 276 | 276 | 0.032 | 0.097 | 0.090 | 0.158 | 0.077 | 0.111 | -0.057 | 0.083 | -0.032 | 0.012 | -0.143 | 0.060 | -0.060 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| eif3b |   | 1 | 116 | 276 | 276 | -0.008 | 0.137 | 0.143 | 0.030 | 0.142 | 0.099 | -0.044 | 0.194 | -0.012 | 0.149 | 0.152 | -0.103 | 0.201 |
|---|
| rps21 |   | 1 | 116 | 276 | 276 | -0.118 | 0.125 | -0.037 | 0.206 | 0.037 | 0.085 | -0.160 | -0.118 | 0.155 | 0.056 | -0.048 | 0.125 | 0.037 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| eif4e |   | 3 | 62 | 260 | 243 | -0.022 | 0.107 | 0.073 | 0.165 | 0.095 | 0.122 | -0.091 | 0.012 | 0.152 | 0.113 | 0.058 | 0.107 | 0.083 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| top2a |   | 15 | 11 | 163 | 134 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.147 | 0.234 | 0.273 |
|---|
GO Enrichment output for subnetwork 6227-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| translational initiation | GO:0006413 |  | 4.759E-08 | 1.095E-04 |
|---|
| regulation of translation | GO:0006417 |  | 2.686E-07 | 3.089E-04 |
|---|
| nuclear export | GO:0051168 |  | 6.065E-07 | 4.65E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.766E-07 | 4.466E-04 |
|---|
| gene silencing by RNA | GO:0031047 |  | 9.306E-07 | 4.281E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.429E-06 | 5.478E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.429E-06 | 4.695E-04 |
|---|
| gene silencing | GO:0016458 |  | 7.387E-06 | 2.124E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.958E-05 | 5.004E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.958E-05 | 4.504E-03 |
|---|
| dermatan sulfate proteoglycan biosynthetic process | GO:0050651 |  | 1.958E-05 | 4.094E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| translational initiation | GO:0006413 |  | 1.838E-08 | 4.49E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.998E-07 | 3.661E-04 |
|---|
| nuclear export | GO:0051168 |  | 3.449E-07 | 2.809E-04 |
|---|
| gene silencing by RNA | GO:0031047 |  | 5.891E-07 | 3.598E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 8.384E-07 | 4.096E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 9.856E-07 | 4.013E-04 |
|---|
| gene silencing | GO:0016458 |  | 4.254E-06 | 1.485E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 9.402E-06 | 2.871E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 9.402E-06 | 2.552E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.29E-05 | 3.151E-03 |
|---|
| dermatan sulfate proteoglycan biosynthetic process | GO:0050651 |  | 1.409E-05 | 3.13E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| translational initiation | GO:0006413 |  | 3.15E-08 | 7.58E-05 |
|---|
| nuclear export | GO:0051168 |  | 4.431E-07 | 5.331E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.124E-07 | 4.109E-04 |
|---|
| gene silencing by RNA | GO:0031047 |  | 8.842E-07 | 5.319E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.258E-06 | 6.054E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.479E-06 | 5.931E-04 |
|---|
| gene silencing | GO:0016458 |  | 6.375E-06 | 2.191E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.234E-05 | 3.712E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.234E-05 | 3.3E-03 |
|---|
| dermatan sulfate proteoglycan biosynthetic process | GO:0050651 |  | 1.234E-05 | 2.97E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.234E-05 | 2.7E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| translational initiation | GO:0006413 |  | 1.838E-08 | 4.49E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.998E-07 | 3.661E-04 |
|---|
| nuclear export | GO:0051168 |  | 3.449E-07 | 2.809E-04 |
|---|
| gene silencing by RNA | GO:0031047 |  | 5.891E-07 | 3.598E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 8.384E-07 | 4.096E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 9.856E-07 | 4.013E-04 |
|---|
| gene silencing | GO:0016458 |  | 4.254E-06 | 1.485E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 9.402E-06 | 2.871E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 9.402E-06 | 2.552E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.29E-05 | 3.151E-03 |
|---|
| dermatan sulfate proteoglycan biosynthetic process | GO:0050651 |  | 1.409E-05 | 3.13E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| translational initiation | GO:0006413 |  | 3.15E-08 | 7.58E-05 |
|---|
| nuclear export | GO:0051168 |  | 4.431E-07 | 5.331E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.124E-07 | 4.109E-04 |
|---|
| gene silencing by RNA | GO:0031047 |  | 8.842E-07 | 5.319E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.258E-06 | 6.054E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.479E-06 | 5.931E-04 |
|---|
| gene silencing | GO:0016458 |  | 6.375E-06 | 2.191E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.234E-05 | 3.712E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.234E-05 | 3.3E-03 |
|---|
| dermatan sulfate proteoglycan biosynthetic process | GO:0050651 |  | 1.234E-05 | 2.97E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.234E-05 | 2.7E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.805E-08 | 1.359E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 2.511E-07 | 4.483E-04 |
|---|
| translational initiation | GO:0006413 |  | 5.438E-07 | 6.473E-04 |
|---|
| cytosolic small ribosomal subunit | GO:0022627 |  | 1.119E-06 | 9.992E-04 |
|---|
| nuclear export | GO:0051168 |  | 3.013E-06 | 2.152E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.618E-06 | 2.748E-03 |
|---|
| 3'-phosphoadenosine 5'-phosphosulfate binding | GO:0050656 |  | 5.061E-06 | 2.582E-03 |
|---|
| dermatan sulfate proteoglycan metabolic process | GO:0050655 |  | 5.061E-06 | 2.259E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.061E-06 | 2.008E-03 |
|---|
| chondroitin sulfotransferase activity | GO:0034481 |  | 5.061E-06 | 1.807E-03 |
|---|
| fibroblast growth factor binding | GO:0017134 |  | 5.061E-06 | 1.643E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.38E-08 | 8.681E-05 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.633E-07 | 2.979E-04 |
|---|
| cytosolic small ribosomal subunit | GO:0022627 |  | 1.302E-06 | 1.583E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.676E-06 | 1.529E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.448E-06 | 1.786E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 3.212E-06 | 1.953E-03 |
|---|
| protein N-terminus binding | GO:0047485 |  | 3.425E-06 | 1.785E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 3.763E-06 | 1.716E-03 |
|---|
| fibroblast growth factor binding | GO:0017134 |  | 3.763E-06 | 1.525E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 3.763E-06 | 1.373E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 3.763E-06 | 1.248E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| translational initiation | GO:0006413 |  | 3.15E-08 | 7.58E-05 |
|---|
| nuclear export | GO:0051168 |  | 4.431E-07 | 5.331E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.124E-07 | 4.109E-04 |
|---|
| gene silencing by RNA | GO:0031047 |  | 8.842E-07 | 5.319E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.258E-06 | 6.054E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.479E-06 | 5.931E-04 |
|---|
| gene silencing | GO:0016458 |  | 6.375E-06 | 2.191E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.234E-05 | 3.712E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.234E-05 | 3.3E-03 |
|---|
| dermatan sulfate proteoglycan biosynthetic process | GO:0050651 |  | 1.234E-05 | 2.97E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.234E-05 | 2.7E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| translational initiation | GO:0006413 |  | 4.759E-08 | 1.095E-04 |
|---|
| regulation of translation | GO:0006417 |  | 2.686E-07 | 3.089E-04 |
|---|
| nuclear export | GO:0051168 |  | 6.065E-07 | 4.65E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.766E-07 | 4.466E-04 |
|---|
| gene silencing by RNA | GO:0031047 |  | 9.306E-07 | 4.281E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.429E-06 | 5.478E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.429E-06 | 4.695E-04 |
|---|
| gene silencing | GO:0016458 |  | 7.387E-06 | 2.124E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.958E-05 | 5.004E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.958E-05 | 4.504E-03 |
|---|
| dermatan sulfate proteoglycan biosynthetic process | GO:0050651 |  | 1.958E-05 | 4.094E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| translational initiation | GO:0006413 |  | 4.759E-08 | 1.095E-04 |
|---|
| regulation of translation | GO:0006417 |  | 2.686E-07 | 3.089E-04 |
|---|
| nuclear export | GO:0051168 |  | 6.065E-07 | 4.65E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.766E-07 | 4.466E-04 |
|---|
| gene silencing by RNA | GO:0031047 |  | 9.306E-07 | 4.281E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.429E-06 | 5.478E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.429E-06 | 4.695E-04 |
|---|
| gene silencing | GO:0016458 |  | 7.387E-06 | 2.124E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.958E-05 | 5.004E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.958E-05 | 4.504E-03 |
|---|
| dermatan sulfate proteoglycan biosynthetic process | GO:0050651 |  | 1.958E-05 | 4.094E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 6.638E-07 | 1.223E-03 |
|---|
| translational initiation | GO:0006413 |  | 6.638E-07 | 6.117E-04 |
|---|
| regulation of translation | GO:0006417 |  | 2.9E-06 | 1.782E-03 |
|---|
| posttranscriptional regulation of gene expression | GO:0010608 |  | 1.183E-05 | 5.453E-03 |
|---|
| dermatan sulfate proteoglycan biosynthetic process | GO:0050651 |  | 2.318E-05 | 8.545E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 2.318E-05 | 7.121E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 3.474E-05 | 9.147E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.474E-05 | 8.004E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 3.474E-05 | 7.114E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 3.474E-05 | 6.403E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 4.86E-05 | 8.142E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| translational initiation | GO:0006413 |  | 4.759E-08 | 1.095E-04 |
|---|
| regulation of translation | GO:0006417 |  | 2.686E-07 | 3.089E-04 |
|---|
| nuclear export | GO:0051168 |  | 6.065E-07 | 4.65E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.766E-07 | 4.466E-04 |
|---|
| gene silencing by RNA | GO:0031047 |  | 9.306E-07 | 4.281E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.429E-06 | 5.478E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.429E-06 | 4.695E-04 |
|---|
| gene silencing | GO:0016458 |  | 7.387E-06 | 2.124E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.958E-05 | 5.004E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.958E-05 | 4.504E-03 |
|---|
| dermatan sulfate proteoglycan biosynthetic process | GO:0050651 |  | 1.958E-05 | 4.094E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| translational initiation | GO:0006413 |  | 3.895E-08 | 9.242E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.454E-07 | 6.471E-04 |
|---|
| gene silencing by RNA | GO:0031047 |  | 6.329E-07 | 5.006E-04 |
|---|
| nuclear export | GO:0051168 |  | 7.332E-07 | 4.35E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.179E-06 | 5.597E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.972E-06 | 7.8E-04 |
|---|
| gene silencing | GO:0016458 |  | 5.63E-06 | 1.909E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.514E-05 | 4.489E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.514E-05 | 3.991E-03 |
|---|
| dermatan sulfate proteoglycan biosynthetic process | GO:0050651 |  | 1.514E-05 | 3.592E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.514E-05 | 3.265E-03 |
|---|



