Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 60598-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2538 | 1.500e-02 | 1.186e-02 | 1.198e-01 |
|---|
| IPC-NIBC-129 | 0.2677 | 4.516e-02 | 6.529e-02 | 4.171e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2301 | 9.530e-03 | 1.592e-02 | 2.653e-01 |
|---|
| Loi_GPL570 | 0.3448 | 1.489e-02 | 1.746e-02 | 7.836e-02 |
|---|
| Loi_GPL96-GPL97 | 0.1576 | 1.000e-06 | 1.000e-05 | 1.000e-06 |
|---|
| Parker_GPL1390 | 0.2420 | 4.356e-02 | 5.447e-02 | 3.650e-01 |
|---|
| Parker_GPL887 | 0.1453 | 2.572e-01 | 2.743e-01 | 2.657e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3968 | 3.563e-01 | 2.838e-01 | 9.074e-01 |
|---|
| Schmidt | 0.2526 | 6.902e-03 | 7.374e-03 | 1.085e-01 |
|---|
| Sotiriou | 0.3322 | 8.684e-03 | 6.615e-03 | 1.737e-01 |
|---|
| Van-De-Vijver | 0.2644 | 7.440e-04 | 1.071e-03 | 4.808e-02 |
|---|
| Zhang | 0.3349 | 1.391e-01 | 9.211e-02 | 2.892e-01 |
|---|
| Zhou | 0.4369 | 4.740e-02 | 3.522e-02 | 3.016e-01 |
|---|
Expression data for subnetwork 60598-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
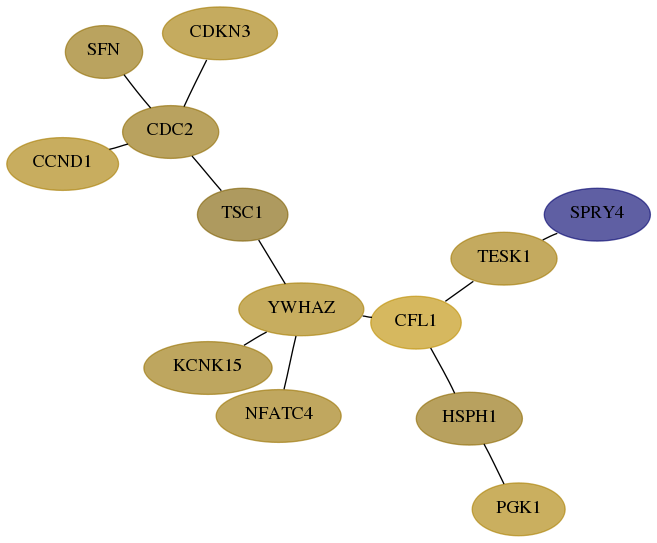
Score for each gene in subnetwork 60598-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| kcnk15 |   | 4 | 50 | 1 | 10 | 0.088 | -0.091 | -0.068 | 0.220 | 0.076 | -0.136 | 0.026 | 0.083 | 0.037 | 0.154 | undef | 0.098 | 0.140 |
|---|
| nfatc4 |   | 15 | 11 | 1 | 5 | 0.094 | -0.056 | -0.021 | 0.204 | 0.079 | -0.008 | 0.243 | 0.060 | 0.077 | -0.012 | 0.136 | 0.131 | 0.154 |
|---|
| ywhaz |   | 17 | 10 | 29 | 25 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | undef | 0.103 | 0.213 |
|---|
| pgk1 |   | 5 | 40 | 18 | 23 | 0.141 | 0.293 | 0.135 | 0.197 | 0.170 | 0.170 | 0.120 | 0.310 | 0.201 | 0.127 | -0.046 | 0.054 | 0.141 |
|---|
| cfl1 |   | 8 | 26 | 18 | 16 | 0.216 | 0.134 | 0.083 | -0.084 | 0.142 | 0.251 | -0.466 | 0.229 | 0.174 | 0.188 | 0.208 | -0.025 | 0.215 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| hsph1 |   | 8 | 26 | 18 | 16 | 0.071 | 0.260 | 0.138 | 0.050 | 0.148 | 0.197 | -0.116 | 0.161 | 0.143 | 0.132 | -0.006 | 0.094 | 0.357 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| tesk1 |   | 7 | 30 | 18 | 19 | 0.110 | 0.169 | 0.161 | 0.179 | 0.096 | 0.138 | -0.154 | 0.245 | 0.155 | 0.092 | 0.067 | 0.029 | 0.200 |
|---|
| spry4 |   | 5 | 40 | 18 | 23 | -0.033 | 0.158 | -0.001 | -0.039 | 0.103 | 0.074 | -0.328 | 0.022 | 0.197 | 0.156 | undef | 0.184 | 0.211 |
|---|
GO Enrichment output for subnetwork 60598-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 9.564E-10 | 2.2E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 2.398E-09 | 2.758E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 2.269E-08 | 1.74E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 2.269E-08 | 1.305E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.899E-07 | 8.737E-05 |
|---|
| glycolysis | GO:0006096 |  | 3.574E-07 | 1.37E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.724E-07 | 1.224E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.538E-07 | 1.88E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 8.251E-07 | 2.109E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.258E-06 | 2.894E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.643E-06 | 3.434E-04 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.554E-10 | 3.795E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 6.62E-09 | 8.086E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 6.62E-09 | 5.391E-06 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 6.62E-09 | 4.043E-06 |
|---|
| neural plate development | GO:0001840 |  | 1.323E-08 | 6.465E-06 |
|---|
| glycolysis | GO:0006096 |  | 8.594E-08 | 3.499E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.088E-07 | 3.798E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.45E-07 | 4.428E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.884E-07 | 5.114E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.922E-07 | 4.696E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.066E-07 | 4.588E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.281E-10 | 3.081E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 6.418E-09 | 7.72E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 6.418E-09 | 5.147E-06 |
|---|
| glycolysis | GO:0006096 |  | 7.441E-08 | 4.476E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.055E-07 | 5.077E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.693E-07 | 6.788E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.827E-07 | 6.278E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.958E-07 | 5.889E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.57E-07 | 9.544E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.777E-07 | 9.087E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 6.287E-07 | 1.375E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.554E-10 | 3.795E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 6.62E-09 | 8.086E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 6.62E-09 | 5.391E-06 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 6.62E-09 | 4.043E-06 |
|---|
| neural plate development | GO:0001840 |  | 1.323E-08 | 6.465E-06 |
|---|
| glycolysis | GO:0006096 |  | 8.594E-08 | 3.499E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.088E-07 | 3.798E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.45E-07 | 4.428E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.884E-07 | 5.114E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.922E-07 | 4.696E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.066E-07 | 4.588E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.281E-10 | 3.081E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 6.418E-09 | 7.72E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 6.418E-09 | 5.147E-06 |
|---|
| glycolysis | GO:0006096 |  | 7.441E-08 | 4.476E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.055E-07 | 5.077E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.693E-07 | 6.788E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.827E-07 | 6.278E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.958E-07 | 5.889E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.57E-07 | 9.544E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.777E-07 | 9.087E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 6.287E-07 | 1.375E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.805E-08 | 1.359E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.618E-06 | 8.245E-03 |
|---|
| neural plate development | GO:0001840 |  | 5.061E-06 | 6.024E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.061E-06 | 4.518E-03 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 5.061E-06 | 3.615E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.589E-06 | 4.517E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.062E-05 | 5.418E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.062E-05 | 4.74E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.088E-05 | 4.317E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.143E-05 | 4.082E-03 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.258E-05 | 4.084E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.43E-08 | 1.251E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.177E-06 | 5.795E-03 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 4.447E-06 | 5.407E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 4.447E-06 | 4.055E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 4.447E-06 | 3.244E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.667E-06 | 4.054E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.667E-06 | 3.475E-03 |
|---|
| neural plate development | GO:0001840 |  | 6.667E-06 | 3.04E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.667E-06 | 2.702E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.064E-05 | 3.88E-03 |
|---|
| regulation of actin cytoskeleton organization | GO:0032956 |  | 1.064E-05 | 3.528E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.281E-10 | 3.081E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 6.418E-09 | 7.72E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 6.418E-09 | 5.147E-06 |
|---|
| glycolysis | GO:0006096 |  | 7.441E-08 | 4.476E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.055E-07 | 5.077E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.693E-07 | 6.788E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.827E-07 | 6.278E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.958E-07 | 5.889E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.57E-07 | 9.544E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.777E-07 | 9.087E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 6.287E-07 | 1.375E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 9.564E-10 | 2.2E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 2.398E-09 | 2.758E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 2.269E-08 | 1.74E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 2.269E-08 | 1.305E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.899E-07 | 8.737E-05 |
|---|
| glycolysis | GO:0006096 |  | 3.574E-07 | 1.37E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.724E-07 | 1.224E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.538E-07 | 1.88E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 8.251E-07 | 2.109E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.258E-06 | 2.894E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.643E-06 | 3.434E-04 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 9.564E-10 | 2.2E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 2.398E-09 | 2.758E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 2.269E-08 | 1.74E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 2.269E-08 | 1.305E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.899E-07 | 8.737E-05 |
|---|
| glycolysis | GO:0006096 |  | 3.574E-07 | 1.37E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.724E-07 | 1.224E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.538E-07 | 1.88E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 8.251E-07 | 2.109E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.258E-06 | 2.894E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.643E-06 | 3.434E-04 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.181E-05 | 0.02176916 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.181E-05 | 0.01088458 |
|---|
| regulation of actin cytoskeleton organization | GO:0032956 |  | 1.431E-05 | 8.794E-03 |
|---|
| regulation of actin filament-based process | GO:0032970 |  | 1.53E-05 | 7.052E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.771E-05 | 6.527E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.771E-05 | 5.439E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 1.771E-05 | 4.662E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 1.771E-05 | 4.079E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 2.478E-05 | 5.074E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.478E-05 | 4.566E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 3.302E-05 | 5.532E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 9.564E-10 | 2.2E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 2.398E-09 | 2.758E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 2.269E-08 | 1.74E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 2.269E-08 | 1.305E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.899E-07 | 8.737E-05 |
|---|
| glycolysis | GO:0006096 |  | 3.574E-07 | 1.37E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.724E-07 | 1.224E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.538E-07 | 1.88E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 8.251E-07 | 2.109E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.258E-06 | 2.894E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.643E-06 | 3.434E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.176E-09 | 2.791E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 2.947E-09 | 3.496E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.98E-08 | 1.566E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.98E-08 | 1.175E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.366E-07 | 1.123E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.33E-07 | 1.712E-04 |
|---|
| glycolysis | GO:0006096 |  | 4.671E-07 | 1.583E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.433E-07 | 1.908E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.065E-06 | 2.807E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.332E-06 | 3.162E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.984E-06 | 4.28E-04 |
|---|



