Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 55605-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2556 | 5.862e-03 | 4.081e-03 | 5.658e-02 |
|---|
| IPC-NIBC-129 | 0.2663 | 3.496e-02 | 5.108e-02 | 3.513e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2757 | 3.373e-03 | 6.158e-03 | 1.440e-01 |
|---|
| Loi_GPL570 | 0.2688 | 4.483e-02 | 5.116e-02 | 2.002e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1282 | 1.000e-06 | 1.100e-05 | 1.000e-06 |
|---|
| Parker_GPL1390 | 0.2896 | 3.334e-02 | 4.220e-02 | 3.133e-01 |
|---|
| Parker_GPL887 | 0.2121 | 2.543e-01 | 2.713e-01 | 2.622e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3557 | 1.814e-01 | 1.264e-01 | 7.223e-01 |
|---|
| Schmidt | 0.2711 | 1.725e-02 | 1.975e-02 | 2.113e-01 |
|---|
| Sotiriou | 0.3205 | 1.056e-02 | 8.088e-03 | 1.943e-01 |
|---|
| Van-De-Vijver | 0.2830 | 7.360e-04 | 1.059e-03 | 4.772e-02 |
|---|
| Zhang | 0.2797 | 2.152e-01 | 1.591e-01 | 4.098e-01 |
|---|
| Zhou | 0.4375 | 6.915e-02 | 5.775e-02 | 4.030e-01 |
|---|
Expression data for subnetwork 55605-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
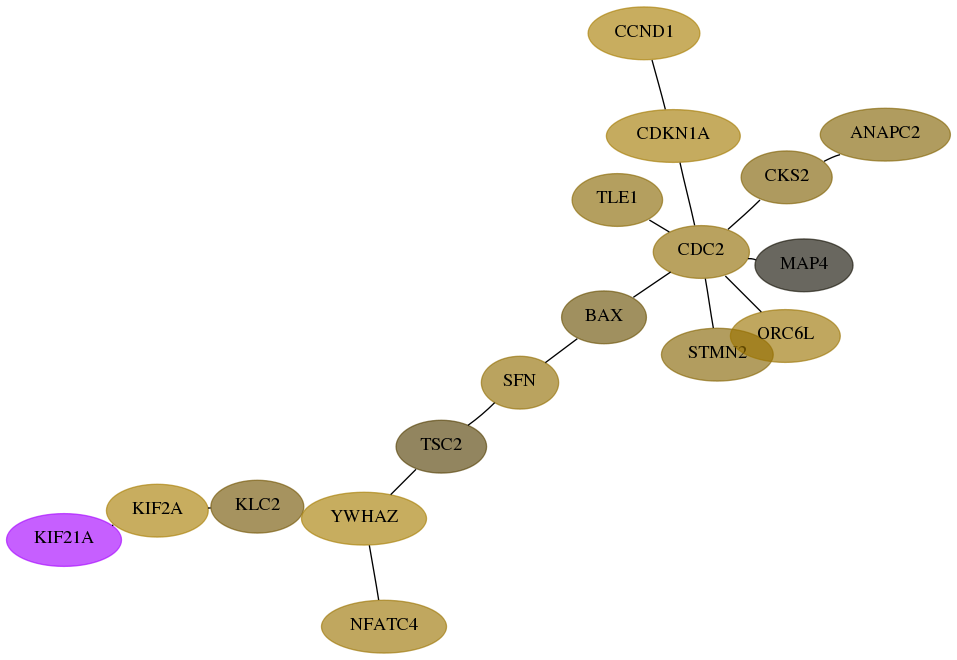
Score for each gene in subnetwork 55605-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| klc2 |   | 2 | 78 | 34 | 42 | 0.036 | -0.030 | 0.148 | 0.074 | 0.099 | 0.019 | 0.066 | 0.141 | 0.025 | 0.198 | undef | 0.098 | 0.185 |
|---|
| kif21a |   | 1 | 116 | 34 | 53 | undef | 0.104 | 0.175 | 0.157 | 0.188 | 0.142 | 0.044 | 0.250 | undef | undef | undef | undef | undef |
|---|
| kif2a |   | 1 | 116 | 34 | 53 | 0.127 | 0.198 | 0.170 | 0.047 | 0.106 | 0.117 | 0.041 | 0.167 | 0.083 | 0.112 | 0.061 | -0.119 | 0.177 |
|---|
| nfatc4 |   | 15 | 11 | 1 | 5 | 0.094 | -0.056 | -0.021 | 0.204 | 0.079 | -0.008 | 0.243 | 0.060 | 0.077 | -0.012 | 0.136 | 0.131 | 0.154 |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| ywhaz |   | 17 | 10 | 29 | 25 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | undef | 0.103 | 0.213 |
|---|
| bax |   | 3 | 62 | 1 | 14 | 0.029 | 0.185 | 0.233 | 0.192 | 0.088 | -0.043 | 0.184 | 0.172 | 0.175 | 0.019 | -0.029 | 0.041 | 0.235 |
|---|
| tsc2 |   | 1 | 116 | 34 | 53 | 0.017 | -0.127 | -0.027 | 0.014 | 0.064 | undef | 0.148 | -0.066 | -0.005 | 0.117 | 0.125 | 0.157 | 0.318 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| cks2 |   | 12 | 18 | 34 | 32 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.080 | 0.284 | 0.383 |
|---|
| orc6l |   | 15 | 11 | 34 | 28 | 0.094 | 0.270 | 0.084 | 0.114 | 0.106 | 0.240 | 0.256 | 0.234 | 0.192 | 0.105 | 0.258 | 0.175 | 0.160 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| map4 |   | 6 | 35 | 18 | 20 | 0.002 | 0.046 | 0.067 | -0.043 | 0.034 | 0.119 | 0.159 | -0.058 | -0.020 | 0.043 | 0.054 | -0.074 | 0.302 |
|---|
| anapc2 |   | 2 | 78 | 34 | 42 | 0.054 | -0.106 | 0.034 | -0.056 | 0.063 | -0.039 | 0.426 | 0.009 | -0.039 | -0.011 | 0.129 | 0.074 | 0.213 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
GO Enrichment output for subnetwork 55605-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.744E-10 | 1.091E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 6.112E-08 | 7.029E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.001E-06 | 7.673E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.375E-06 | 1.94E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.872E-06 | 1.781E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.154E-06 | 1.592E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 5.089E-06 | 1.672E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 6.835E-06 | 1.965E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.207E-05 | 3.085E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.165E-05 | 4.981E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.928E-05 | 6.123E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.583E-11 | 1.364E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.422E-08 | 1.737E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.041E-07 | 3.291E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.894E-07 | 4.821E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.247E-06 | 6.094E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.322E-06 | 5.382E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.653E-06 | 5.77E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.636E-06 | 1.11E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.574E-06 | 1.242E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 6.903E-06 | 1.686E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 9.905E-06 | 2.2E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.28E-10 | 3.079E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 2.166E-08 | 2.606E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 6.15E-07 | 4.932E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.201E-06 | 7.223E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.924E-06 | 9.26E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.043E-06 | 8.191E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.571E-06 | 8.835E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.308E-06 | 1.296E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.891E-06 | 1.308E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.048E-05 | 2.522E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.503E-05 | 3.288E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.583E-11 | 1.364E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.422E-08 | 1.737E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.041E-07 | 3.291E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.894E-07 | 4.821E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.247E-06 | 6.094E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.322E-06 | 5.382E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.653E-06 | 5.77E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.636E-06 | 1.11E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.574E-06 | 1.242E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 6.903E-06 | 1.686E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 9.905E-06 | 2.2E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.28E-10 | 3.079E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 2.166E-08 | 2.606E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 6.15E-07 | 4.932E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.201E-06 | 7.223E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.924E-06 | 9.26E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.043E-06 | 8.191E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.571E-06 | 8.835E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.308E-06 | 1.296E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.891E-06 | 1.308E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.048E-05 | 2.522E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.503E-05 | 3.288E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.475E-12 | 1.955E-08 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 2.285E-10 | 4.08E-07 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 3.829E-09 | 4.558E-06 |
|---|
| kinase regulator activity | GO:0019207 |  | 9.85E-09 | 8.794E-06 |
|---|
| microtubule associated complex | GO:0005875 |  | 3.177E-08 | 2.269E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 4.766E-08 | 2.837E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 3.087E-07 | 1.575E-04 |
|---|
| microtubule motor activity | GO:0003777 |  | 3.999E-07 | 1.785E-04 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 2.754E-06 | 1.093E-03 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 3.409E-06 | 1.217E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.028E-05 | 3.337E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.461E-12 | 1.992E-08 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.952E-10 | 1.086E-06 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 3.86E-09 | 4.694E-06 |
|---|
| kinase regulator activity | GO:0019207 |  | 7.934E-09 | 7.235E-06 |
|---|
| microtubule associated complex | GO:0005875 |  | 2.734E-08 | 1.995E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 4.168E-08 | 2.534E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 5.925E-08 | 3.088E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.699E-07 | 1.231E-04 |
|---|
| microtubule motor activity | GO:0003777 |  | 3.223E-07 | 1.306E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.667E-07 | 1.338E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 3.905E-07 | 1.295E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.28E-10 | 3.079E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 2.166E-08 | 2.606E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 6.15E-07 | 4.932E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.201E-06 | 7.223E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.924E-06 | 9.26E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.043E-06 | 8.191E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.571E-06 | 8.835E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.308E-06 | 1.296E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.891E-06 | 1.308E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.048E-05 | 2.522E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.503E-05 | 3.288E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.744E-10 | 1.091E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 6.112E-08 | 7.029E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.001E-06 | 7.673E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.375E-06 | 1.94E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.872E-06 | 1.781E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.154E-06 | 1.592E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 5.089E-06 | 1.672E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 6.835E-06 | 1.965E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.207E-05 | 3.085E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.165E-05 | 4.981E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.928E-05 | 6.123E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.744E-10 | 1.091E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 6.112E-08 | 7.029E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.001E-06 | 7.673E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.375E-06 | 1.94E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.872E-06 | 1.781E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.154E-06 | 1.592E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 5.089E-06 | 1.672E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 6.835E-06 | 1.965E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.207E-05 | 3.085E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.165E-05 | 4.981E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.928E-05 | 6.123E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.7E-07 | 4.975E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.781E-05 | 0.02562666 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 2.781E-05 | 0.01708444 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 2.781E-05 | 0.01281333 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.047E-05 | 0.01123212 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 3.305E-05 | 0.0101524 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 3.863E-05 | 0.01017157 |
|---|
| regulation of caspase activity | GO:0043281 |  | 4.164E-05 | 9.593E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 4.167E-05 | 8.534E-03 |
|---|
| regulation of phosphoinositide 3-kinase cascade | GO:0014066 |  | 4.167E-05 | 7.681E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.48E-05 | 7.506E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.744E-10 | 1.091E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 6.112E-08 | 7.029E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.001E-06 | 7.673E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.375E-06 | 1.94E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.872E-06 | 1.781E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.154E-06 | 1.592E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 5.089E-06 | 1.672E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 6.835E-06 | 1.965E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.207E-05 | 3.085E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.165E-05 | 4.981E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.928E-05 | 6.123E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.836E-10 | 1.385E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 5.912E-08 | 7.015E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.289E-06 | 1.02E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.959E-06 | 2.349E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 5.235E-06 | 2.484E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 5.582E-06 | 2.208E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 6.727E-06 | 2.28E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 8.908E-06 | 2.642E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.496E-05 | 3.945E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.57E-05 | 6.1E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 3.409E-05 | 7.354E-03 |
|---|



