Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 51022-5-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2190 | 5.443e-03 | 3.750e-03 | 5.323e-02 |
|---|
| IPC-NIBC-129 | 0.2606 | 8.260e-02 | 1.159e-01 | 5.939e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2793 | 1.334e-02 | 2.165e-02 | 3.182e-01 |
|---|
| Loi_GPL570 | 0.2593 | 5.130e-02 | 5.833e-02 | 2.230e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1770 | 1.000e-06 | 1.600e-05 | 2.000e-06 |
|---|
| Parker_GPL1390 | 0.2265 | 4.290e-02 | 5.368e-02 | 3.619e-01 |
|---|
| Parker_GPL887 | 0.0911 | 3.198e-01 | 3.381e-01 | 3.428e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3497 | 5.844e-01 | 5.160e-01 | 9.834e-01 |
|---|
| Schmidt | 0.2536 | 3.117e-02 | 3.709e-02 | 3.128e-01 |
|---|
| Sotiriou | 0.3486 | 6.596e-03 | 4.985e-03 | 1.479e-01 |
|---|
| Van-De-Vijver | 0.2199 | 1.602e-03 | 2.304e-03 | 8.013e-02 |
|---|
| Zhang | 0.2436 | 1.038e-01 | 6.370e-02 | 2.265e-01 |
|---|
| Zhou | 0.3940 | 7.305e-02 | 6.201e-02 | 4.193e-01 |
|---|
Expression data for subnetwork 51022-5-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
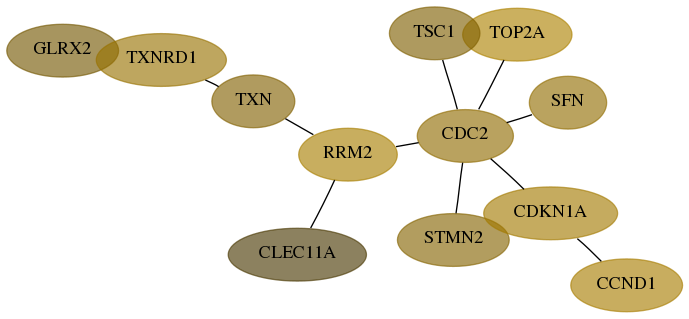
Score for each gene in subnetwork 51022-5-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| rrm2 |   | 5 | 40 | 163 | 143 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | undef | 0.129 | 0.302 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| txn |   | 1 | 116 | 163 | 170 | 0.052 | 0.168 | 0.117 | 0.209 | 0.171 | 0.155 | 0.166 | 0.246 | 0.161 | 0.168 | 0.057 | 0.132 | 0.324 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| top2a |   | 15 | 11 | 163 | 134 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.147 | 0.234 | 0.273 |
|---|
| glrx2 |   | 1 | 116 | 163 | 170 | 0.038 | 0.231 | 0.122 | 0.021 | 0.152 | -0.160 | 0.135 | 0.284 | 0.131 | 0.171 | 0.132 | 0.070 | 0.130 |
|---|
| txnrd1 |   | 1 | 116 | 163 | 170 | 0.090 | 0.133 | 0.124 | 0.256 | 0.156 | 0.162 | 0.204 | 0.101 | 0.169 | 0.142 | undef | 0.118 | 0.215 |
|---|
| clec11a |   | 2 | 78 | 163 | 156 | 0.013 | 0.101 | 0.071 | -0.085 | 0.066 | 0.082 | 0.209 | -0.126 | -0.034 | -0.001 | -0.039 | 0.059 | -0.075 |
|---|
GO Enrichment output for subnetwork 51022-5-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.555E-08 | 5.877E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.468E-08 | 5.139E-05 |
|---|
| cell redox homeostasis | GO:0045454 |  | 1.971E-07 | 1.511E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.007E-07 | 1.729E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 8.063E-07 | 3.709E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.681E-06 | 6.443E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.681E-06 | 5.522E-04 |
|---|
| electron transport chain | GO:0022900 |  | 3.555E-06 | 1.022E-03 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 4.591E-06 | 1.173E-03 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 8.925E-06 | 2.053E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.231E-05 | 2.575E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell redox homeostasis | GO:0045454 |  | 4.605E-12 | 1.125E-08 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 1.957E-11 | 2.39E-08 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 7.605E-11 | 6.193E-08 |
|---|
| cellular response to hydrogen peroxide | GO:0070301 |  | 5.129E-10 | 3.132E-07 |
|---|
| cellular response to reactive oxygen species | GO:0034614 |  | 1.17E-09 | 5.715E-07 |
|---|
| hydrogen peroxide metabolic process | GO:0042743 |  | 1.972E-09 | 8.03E-07 |
|---|
| cellular response to oxidative stress | GO:0034599 |  | 2.314E-09 | 8.077E-07 |
|---|
| mesoderm formation | GO:0001707 |  | 4.732E-09 | 1.445E-06 |
|---|
| formation of primary germ layer | GO:0001704 |  | 6.097E-09 | 1.655E-06 |
|---|
| mesoderm morphogenesis | GO:0048332 |  | 6.881E-09 | 1.681E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.142E-08 | 1.142E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell redox homeostasis | GO:0045454 |  | 1.207E-09 | 2.904E-06 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 9.16E-09 | 1.102E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.359E-08 | 1.09E-05 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 2.837E-08 | 1.706E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 3.258E-08 | 1.568E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.936E-08 | 3.984E-05 |
|---|
| cellular response to hydrogen peroxide | GO:0070301 |  | 3.151E-07 | 1.083E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 3.55E-07 | 1.068E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 5.935E-07 | 1.587E-04 |
|---|
| cellular response to reactive oxygen species | GO:0034614 |  | 5.935E-07 | 1.428E-04 |
|---|
| hydrogen peroxide metabolic process | GO:0042743 |  | 8.85E-07 | 1.936E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell redox homeostasis | GO:0045454 |  | 4.605E-12 | 1.125E-08 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 1.957E-11 | 2.39E-08 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 7.605E-11 | 6.193E-08 |
|---|
| cellular response to hydrogen peroxide | GO:0070301 |  | 5.129E-10 | 3.132E-07 |
|---|
| cellular response to reactive oxygen species | GO:0034614 |  | 1.17E-09 | 5.715E-07 |
|---|
| hydrogen peroxide metabolic process | GO:0042743 |  | 1.972E-09 | 8.03E-07 |
|---|
| cellular response to oxidative stress | GO:0034599 |  | 2.314E-09 | 8.077E-07 |
|---|
| mesoderm formation | GO:0001707 |  | 4.732E-09 | 1.445E-06 |
|---|
| formation of primary germ layer | GO:0001704 |  | 6.097E-09 | 1.655E-06 |
|---|
| mesoderm morphogenesis | GO:0048332 |  | 6.881E-09 | 1.681E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.142E-08 | 1.142E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell redox homeostasis | GO:0045454 |  | 1.207E-09 | 2.904E-06 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 9.16E-09 | 1.102E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.359E-08 | 1.09E-05 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 2.837E-08 | 1.706E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 3.258E-08 | 1.568E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.936E-08 | 3.984E-05 |
|---|
| cellular response to hydrogen peroxide | GO:0070301 |  | 3.151E-07 | 1.083E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 3.55E-07 | 1.068E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 5.935E-07 | 1.587E-04 |
|---|
| cellular response to reactive oxygen species | GO:0034614 |  | 5.935E-07 | 1.428E-04 |
|---|
| hydrogen peroxide metabolic process | GO:0042743 |  | 8.85E-07 | 1.936E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.73E-08 | 6.178E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.805E-08 | 6.793E-05 |
|---|
| cell redox homeostasis | GO:0045454 |  | 3.805E-08 | 4.529E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.091E-08 | 4.545E-05 |
|---|
| protein disulfide oxidoreductase activity | GO:0015035 |  | 6.786E-08 | 4.846E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.044E-07 | 6.214E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.122E-07 | 5.723E-05 |
|---|
| disulfide oxidoreductase activity | GO:0015036 |  | 1.402E-07 | 6.257E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 2.213E-07 | 8.78E-05 |
|---|
| electron transport chain | GO:0022900 |  | 6.356E-07 | 2.27E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 7.974E-07 | 2.589E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.462E-08 | 5.334E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.43E-08 | 6.257E-05 |
|---|
| cell redox homeostasis | GO:0045454 |  | 3.715E-08 | 4.517E-05 |
|---|
| protein disulfide oxidoreductase activity | GO:0015035 |  | 4.303E-08 | 3.924E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 9.481E-08 | 6.917E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 9.836E-08 | 5.98E-05 |
|---|
| disulfide oxidoreductase activity | GO:0015036 |  | 1.185E-07 | 6.174E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.457E-07 | 6.646E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.744E-07 | 7.07E-05 |
|---|
| electron transport chain | GO:0022900 |  | 4.673E-07 | 1.705E-04 |
|---|
| oxidoreductase activity. acting on sulfur group of donors | GO:0016667 |  | 1.41E-06 | 4.677E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell redox homeostasis | GO:0045454 |  | 1.207E-09 | 2.904E-06 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 9.16E-09 | 1.102E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.359E-08 | 1.09E-05 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 2.837E-08 | 1.706E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 3.258E-08 | 1.568E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.936E-08 | 3.984E-05 |
|---|
| cellular response to hydrogen peroxide | GO:0070301 |  | 3.151E-07 | 1.083E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 3.55E-07 | 1.068E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 5.935E-07 | 1.587E-04 |
|---|
| cellular response to reactive oxygen species | GO:0034614 |  | 5.935E-07 | 1.428E-04 |
|---|
| hydrogen peroxide metabolic process | GO:0042743 |  | 8.85E-07 | 1.936E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.555E-08 | 5.877E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.468E-08 | 5.139E-05 |
|---|
| cell redox homeostasis | GO:0045454 |  | 1.971E-07 | 1.511E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.007E-07 | 1.729E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 8.063E-07 | 3.709E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.681E-06 | 6.443E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.681E-06 | 5.522E-04 |
|---|
| electron transport chain | GO:0022900 |  | 3.555E-06 | 1.022E-03 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 4.591E-06 | 1.173E-03 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 8.925E-06 | 2.053E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.231E-05 | 2.575E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.555E-08 | 5.877E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.468E-08 | 5.139E-05 |
|---|
| cell redox homeostasis | GO:0045454 |  | 1.971E-07 | 1.511E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.007E-07 | 1.729E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 8.063E-07 | 3.709E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.681E-06 | 6.443E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.681E-06 | 5.522E-04 |
|---|
| electron transport chain | GO:0022900 |  | 3.555E-06 | 1.022E-03 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 4.591E-06 | 1.173E-03 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 8.925E-06 | 2.053E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.231E-05 | 2.575E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell redox homeostasis | GO:0045454 |  | 5.039E-06 | 9.287E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.181E-05 | 0.01088458 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.771E-05 | 0.01087825 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.771E-05 | 8.159E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 1.771E-05 | 6.527E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 1.771E-05 | 5.439E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 2.478E-05 | 6.523E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.478E-05 | 5.708E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 3.302E-05 | 6.761E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 3.302E-05 | 6.085E-03 |
|---|
| positive regulation of cell size | GO:0045793 |  | 4.242E-05 | 7.108E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.555E-08 | 5.877E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.468E-08 | 5.139E-05 |
|---|
| cell redox homeostasis | GO:0045454 |  | 1.971E-07 | 1.511E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.007E-07 | 1.729E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 8.063E-07 | 3.709E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.681E-06 | 6.443E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.681E-06 | 5.522E-04 |
|---|
| electron transport chain | GO:0022900 |  | 3.555E-06 | 1.022E-03 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 4.591E-06 | 1.173E-03 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 8.925E-06 | 2.053E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.231E-05 | 2.575E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.714E-08 | 1.119E-04 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.537E-08 | 8.943E-05 |
|---|
| cell redox homeostasis | GO:0045454 |  | 3.021E-07 | 2.39E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.833E-07 | 2.274E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 9.34E-07 | 4.433E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.051E-06 | 8.112E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 2.357E-06 | 7.99E-04 |
|---|
| electron transport chain | GO:0022900 |  | 4.757E-06 | 1.411E-03 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 5.946E-06 | 1.568E-03 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 1.202E-05 | 2.851E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.277E-05 | 2.754E-03 |
|---|



