Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 3678-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2343 | 1.653e-02 | 1.323e-02 | 1.292e-01 |
|---|
| IPC-NIBC-129 | 0.2984 | 1.955e-02 | 2.917e-02 | 2.285e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2254 | 1.232e-02 | 2.013e-02 | 3.050e-01 |
|---|
| Loi_GPL570 | 0.1508 | 2.261e-01 | 2.458e-01 | 6.248e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1894 | 0.000e+00 | 1.000e-06 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2302 | 9.950e-02 | 1.195e-01 | 5.594e-01 |
|---|
| Parker_GPL887 | 0.1781 | 2.908e-01 | 3.086e-01 | 3.070e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3540 | 2.576e-01 | 1.923e-01 | 8.264e-01 |
|---|
| Schmidt | 0.1944 | 3.485e-02 | 4.173e-02 | 3.354e-01 |
|---|
| Sotiriou | 0.2952 | 1.610e-02 | 1.249e-02 | 2.460e-01 |
|---|
| Van-De-Vijver | 0.3248 | 1.100e-04 | 1.570e-04 | 1.238e-02 |
|---|
| Zhang | 0.2368 | 8.591e-02 | 5.019e-02 | 1.928e-01 |
|---|
| Zhou | 0.5432 | 7.022e-02 | 5.891e-02 | 4.076e-01 |
|---|
Expression data for subnetwork 3678-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
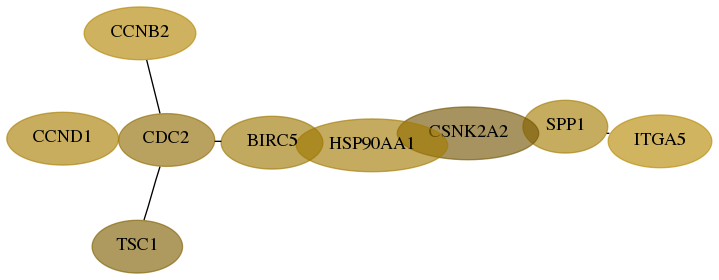
Score for each gene in subnetwork 3678-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| ccnb2 |   | 12 | 18 | 84 | 62 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.178 | 0.090 | 0.366 | 0.240 | 0.169 | 0.279 | 0.211 | 0.463 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| spp1 |   | 3 | 62 | 106 | 95 | 0.121 | 0.316 | 0.018 | -0.074 | 0.132 | 0.124 | -0.237 | 0.185 | 0.031 | 0.109 | 0.212 | 0.141 | 0.275 |
|---|
| csnk2a2 |   | 2 | 78 | 106 | 103 | 0.036 | 0.182 | 0.089 | -0.033 | 0.074 | 0.094 | -0.125 | 0.149 | 0.060 | 0.090 | 0.051 | -0.155 | 0.174 |
|---|
| hsp90aa1 |   | 5 | 40 | 106 | 90 | 0.116 | 0.030 | 0.164 | 0.076 | 0.119 | 0.155 | -0.199 | 0.169 | 0.069 | 0.102 | undef | -0.011 | 0.399 |
|---|
| birc5 |   | 5 | 40 | 103 | 89 | 0.101 | 0.216 | 0.190 | 0.134 | 0.223 | 0.149 | 0.166 | 0.202 | 0.236 | 0.241 | undef | 0.205 | 0.400 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| itga5 |   | 1 | 116 | 106 | 116 | 0.182 | 0.283 | 0.049 | -0.041 | 0.072 | 0.167 | -0.001 | 0.015 | -0.084 | -0.025 | 0.193 | -0.051 | 0.198 |
|---|
GO Enrichment output for subnetwork 3678-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.841E-06 | 0.01803374 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 7.841E-06 | 9.017E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 7.841E-06 | 6.011E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.719E-06 | 5.013E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.175E-05 | 5.407E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.175E-05 | 4.506E-03 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 1.175E-05 | 3.862E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.175E-05 | 3.38E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.645E-05 | 4.203E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.645E-05 | 3.783E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.645E-05 | 3.439E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.859E-06 | 4.542E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.42E-06 | 2.956E-03 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 2.42E-06 | 1.971E-03 |
|---|
| protein refolding | GO:0042026 |  | 3.629E-06 | 2.216E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.629E-06 | 1.773E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 5.079E-06 | 2.068E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 5.079E-06 | 1.773E-03 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 5.079E-06 | 1.551E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.77E-06 | 1.838E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 6.77E-06 | 1.654E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 6.77E-06 | 1.504E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.543E-06 | 6.119E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.996E-06 | 3.604E-03 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 2.996E-06 | 2.403E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.996E-06 | 1.802E-03 |
|---|
| protein refolding | GO:0042026 |  | 4.492E-06 | 2.162E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 6.287E-06 | 2.521E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 6.287E-06 | 2.161E-03 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 6.287E-06 | 1.891E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.38E-06 | 2.24E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 8.38E-06 | 2.016E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 8.38E-06 | 1.833E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.859E-06 | 4.542E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.42E-06 | 2.956E-03 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 2.42E-06 | 1.971E-03 |
|---|
| protein refolding | GO:0042026 |  | 3.629E-06 | 2.216E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.629E-06 | 1.773E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 5.079E-06 | 2.068E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 5.079E-06 | 1.773E-03 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 5.079E-06 | 1.551E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.77E-06 | 1.838E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 6.77E-06 | 1.654E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 6.77E-06 | 1.504E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.543E-06 | 6.119E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.996E-06 | 3.604E-03 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 2.996E-06 | 2.403E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.996E-06 | 1.802E-03 |
|---|
| protein refolding | GO:0042026 |  | 4.492E-06 | 2.162E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 6.287E-06 | 2.521E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 6.287E-06 | 2.161E-03 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 6.287E-06 | 1.891E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.38E-06 | 2.24E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 8.38E-06 | 2.016E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 8.38E-06 | 1.833E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein N-terminus binding | GO:0047485 |  | 1.125E-06 | 4.019E-03 |
|---|
| ruffle membrane | GO:0032587 |  | 1.933E-06 | 3.452E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.933E-06 | 2.302E-03 |
|---|
| nitric-oxide synthase regulator activity | GO:0030235 |  | 1.933E-06 | 1.726E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.933E-06 | 1.381E-03 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 1.933E-06 | 1.151E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.9E-06 | 1.479E-03 |
|---|
| leading edge membrane | GO:0031256 |  | 2.9E-06 | 1.294E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 2.967E-06 | 1.177E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 3.862E-06 | 1.379E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 4.058E-06 | 1.318E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| midbody | GO:0030496 |  | 1.633E-07 | 5.958E-04 |
|---|
| spindle microtubule | GO:0005876 |  | 6.036E-07 | 1.101E-03 |
|---|
| chaperone binding | GO:0051087 |  | 8.423E-07 | 1.024E-03 |
|---|
| ruffle membrane | GO:0032587 |  | 8.717E-06 | 7.95E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 8.717E-06 | 6.36E-03 |
|---|
| nitric-oxide synthase regulator activity | GO:0030235 |  | 8.717E-06 | 5.3E-03 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 8.717E-06 | 4.543E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 8.717E-06 | 3.975E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 8.717E-06 | 3.533E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.993E-06 | 3.281E-03 |
|---|
| protein N-terminus binding | GO:0047485 |  | 1.257E-05 | 4.167E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.543E-06 | 6.119E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.996E-06 | 3.604E-03 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 2.996E-06 | 2.403E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.996E-06 | 1.802E-03 |
|---|
| protein refolding | GO:0042026 |  | 4.492E-06 | 2.162E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 6.287E-06 | 2.521E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 6.287E-06 | 2.161E-03 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 6.287E-06 | 1.891E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.38E-06 | 2.24E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 8.38E-06 | 2.016E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 8.38E-06 | 1.833E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.841E-06 | 0.01803374 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 7.841E-06 | 9.017E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 7.841E-06 | 6.011E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.719E-06 | 5.013E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.175E-05 | 5.407E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.175E-05 | 4.506E-03 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 1.175E-05 | 3.862E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.175E-05 | 3.38E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.645E-05 | 4.203E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.645E-05 | 3.783E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.645E-05 | 3.439E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.841E-06 | 0.01803374 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 7.841E-06 | 9.017E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 7.841E-06 | 6.011E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.719E-06 | 5.013E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.175E-05 | 5.407E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.175E-05 | 4.506E-03 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 1.175E-05 | 3.862E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.175E-05 | 3.38E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.645E-05 | 4.203E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.645E-05 | 3.783E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.645E-05 | 3.439E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| memory | GO:0007613 |  | 6.331E-06 | 0.01166884 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 6.331E-06 | 5.834E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 9.493E-06 | 5.832E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 9.493E-06 | 4.374E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 9.493E-06 | 3.499E-03 |
|---|
| thymus development | GO:0048538 |  | 9.493E-06 | 2.916E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 1.329E-05 | 3.498E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 1.771E-05 | 4.079E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.771E-05 | 3.626E-03 |
|---|
| regulation of dephosphorylation | GO:0035303 |  | 2.276E-05 | 4.194E-03 |
|---|
| regulation of actin filament bundle formation | GO:0032231 |  | 2.276E-05 | 3.813E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.841E-06 | 0.01803374 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 7.841E-06 | 9.017E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 7.841E-06 | 6.011E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.719E-06 | 5.013E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.175E-05 | 5.407E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.175E-05 | 4.506E-03 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 1.175E-05 | 3.862E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.175E-05 | 3.38E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.645E-05 | 4.203E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.645E-05 | 3.783E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.645E-05 | 3.439E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein refolding | GO:0042026 |  | 1.036E-08 | 2.458E-05 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.812E-08 | 2.15E-05 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 2.897E-08 | 2.292E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 5.551E-07 | 3.293E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 5.861E-07 | 2.782E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 7.088E-07 | 2.803E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.038E-06 | 3.52E-04 |
|---|
| mitochondrial membrane organization | GO:0007006 |  | 1.179E-06 | 3.498E-04 |
|---|
| mitochondrial transport | GO:0006839 |  | 6.268E-06 | 1.653E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.867E-06 | 1.629E-03 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 6.867E-06 | 1.481E-03 |
|---|



