Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 292-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2535 | 1.309e-02 | 1.017e-02 | 1.078e-01 |
|---|
| IPC-NIBC-129 | 0.2684 | 3.119e-02 | 4.578e-02 | 3.243e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2368 | 1.151e-02 | 1.892e-02 | 2.941e-01 |
|---|
| Loi_GPL570 | 0.3363 | 1.686e-02 | 1.971e-02 | 8.746e-02 |
|---|
| Loi_GPL96-GPL97 | 0.1458 | 1.000e-06 | 1.000e-05 | 1.000e-06 |
|---|
| Parker_GPL1390 | 0.2333 | 9.222e-02 | 1.112e-01 | 5.397e-01 |
|---|
| Parker_GPL887 | 0.0055 | 2.577e-01 | 2.749e-01 | 2.664e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3710 | 9.940e-01 | 9.925e-01 | 1.000e+00 |
|---|
| Schmidt | 0.1999 | 2.853e-02 | 3.377e-02 | 2.957e-01 |
|---|
| Sotiriou | 0.3303 | 8.970e-03 | 6.840e-03 | 1.770e-01 |
|---|
| Van-De-Vijver | 0.2912 | 1.159e-03 | 1.668e-03 | 6.477e-02 |
|---|
| Zhang | 0.2490 | 1.660e-01 | 1.150e-01 | 3.338e-01 |
|---|
| Zhou | 0.4121 | 6.010e-02 | 4.811e-02 | 3.632e-01 |
|---|
Expression data for subnetwork 292-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
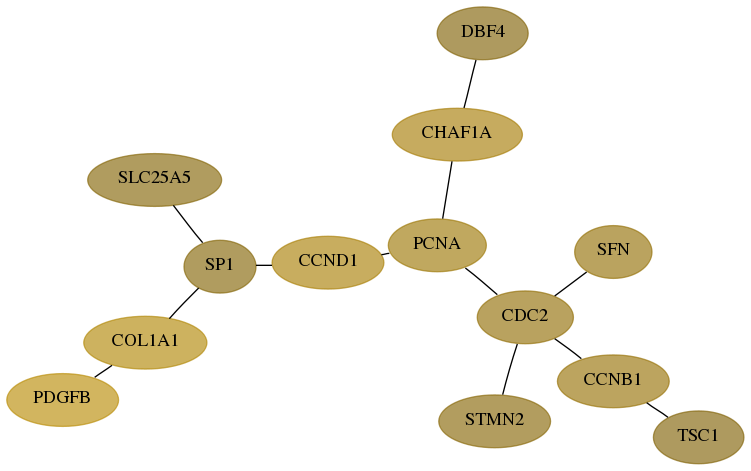
Subnetwork structure for each dataset
- Desmedt
Score for each gene in subnetwork 292-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| pdgfb |   | 3 | 62 | 95 | 86 | 0.186 | -0.069 | 0.089 | 0.269 | 0.087 | 0.107 | -0.031 | 0.163 | 0.038 | 0.139 | 0.143 | 0.037 | 0.170 |
|---|
| pcna |   | 15 | 11 | 110 | 85 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.119 | 0.104 | 0.256 | 0.151 | 0.059 | 0.201 | 0.123 | 0.233 |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| chaf1a |   | 1 | 116 | 142 | 147 | 0.118 | 0.111 | 0.131 | -0.035 | 0.092 | 0.186 | 0.248 | 0.072 | 0.209 | 0.025 | 0.140 | 0.180 | 0.114 |
|---|
| slc25a5 |   | 1 | 116 | 142 | 147 | 0.053 | 0.214 | 0.106 | 0.218 | 0.115 | undef | -0.171 | 0.221 | 0.177 | 0.141 | 0.133 | -0.019 | 0.081 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| dbf4 |   | 1 | 116 | 142 | 147 | 0.050 | 0.253 | 0.072 | 0.170 | 0.110 | 0.165 | 0.165 | 0.207 | 0.139 | 0.129 | 0.089 | 0.138 | 0.223 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| sp1 |   | 8 | 26 | 71 | 57 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.033 | -0.370 | 0.202 | 0.284 | 0.227 | 0.304 | 0.091 | -0.068 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| col1a1 |   | 2 | 78 | 142 | 138 | 0.157 | 0.030 | -0.010 | -0.046 | -0.012 | 0.017 | 0.144 | -0.059 | -0.107 | -0.056 | -0.051 | -0.044 | 0.192 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
GO Enrichment output for subnetwork 292-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 5.775E-07 | 1.328E-03 |
|---|
| regulation of blood coagulation | GO:0030193 |  | 1.945E-06 | 2.236E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.49E-06 | 1.909E-03 |
|---|
| interphase | GO:0051325 |  | 2.731E-06 | 1.57E-03 |
|---|
| regulation of coagulation | GO:0050818 |  | 3.274E-06 | 1.506E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.241E-05 | 4.757E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.241E-05 | 4.077E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 1.241E-05 | 3.568E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.241E-05 | 3.171E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.535E-05 | 3.53E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 3.684E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 2.679E-07 | 6.544E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.621E-07 | 5.644E-04 |
|---|
| regulation of blood coagulation | GO:0030193 |  | 6.093E-07 | 4.962E-04 |
|---|
| regulation of coagulation | GO:0050818 |  | 9.499E-07 | 5.802E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.528E-06 | 1.724E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.001E-06 | 1.629E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.001E-06 | 1.396E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 4.001E-06 | 1.222E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.016E-06 | 1.09E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 5.998E-06 | 1.465E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 5.998E-06 | 1.332E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 3.253E-07 | 7.827E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.505E-07 | 7.826E-04 |
|---|
| interphase | GO:0051325 |  | 7.371E-07 | 5.911E-04 |
|---|
| regulation of blood coagulation | GO:0030193 |  | 7.7E-07 | 4.631E-04 |
|---|
| regulation of coagulation | GO:0050818 |  | 1.221E-06 | 5.875E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.032E-06 | 2.018E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 1.748E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.087E-06 | 1.53E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 5.087E-06 | 1.36E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 1.224E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.087E-06 | 1.113E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 2.679E-07 | 6.544E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.621E-07 | 5.644E-04 |
|---|
| regulation of blood coagulation | GO:0030193 |  | 6.093E-07 | 4.962E-04 |
|---|
| regulation of coagulation | GO:0050818 |  | 9.499E-07 | 5.802E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.528E-06 | 1.724E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.001E-06 | 1.629E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.001E-06 | 1.396E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 4.001E-06 | 1.222E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.016E-06 | 1.09E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 5.998E-06 | 1.465E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 5.998E-06 | 1.332E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 3.253E-07 | 7.827E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.505E-07 | 7.826E-04 |
|---|
| interphase | GO:0051325 |  | 7.371E-07 | 5.911E-04 |
|---|
| regulation of blood coagulation | GO:0030193 |  | 7.7E-07 | 4.631E-04 |
|---|
| regulation of coagulation | GO:0050818 |  | 1.221E-06 | 5.875E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.032E-06 | 2.018E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 1.748E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.087E-06 | 1.53E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 5.087E-06 | 1.36E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 1.224E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.087E-06 | 1.113E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.73E-08 | 6.178E-05 |
|---|
| skin development | GO:0043588 |  | 1.402E-07 | 2.503E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.854E-07 | 3.398E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.618E-06 | 4.122E-03 |
|---|
| embryonic camera-type eye development | GO:0031076 |  | 5.061E-06 | 3.615E-03 |
|---|
| MutLalpha complex binding | GO:0032405 |  | 5.061E-06 | 3.012E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.061E-06 | 2.582E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.061E-06 | 2.259E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.952E-06 | 2.362E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 5.952E-06 | 2.126E-03 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 7.106E-06 | 2.307E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.173E-07 | 4.279E-04 |
|---|
| skin development | GO:0043588 |  | 1.413E-06 | 2.578E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.839E-06 | 4.669E-03 |
|---|
| interphase | GO:0051325 |  | 4.233E-06 | 3.861E-03 |
|---|
| enucleate erythrocyte differentiation | GO:0043353 |  | 1.708E-05 | 0.01246178 |
|---|
| platelet-derived growth factor binding | GO:0048407 |  | 1.708E-05 | 0.01038481 |
|---|
| platelet dense granule | GO:0042827 |  | 1.708E-05 | 8.901E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.708E-05 | 7.789E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 1.708E-05 | 6.923E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 1.708E-05 | 6.231E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.506E-05 | 8.312E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 3.253E-07 | 7.827E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.505E-07 | 7.826E-04 |
|---|
| interphase | GO:0051325 |  | 7.371E-07 | 5.911E-04 |
|---|
| regulation of blood coagulation | GO:0030193 |  | 7.7E-07 | 4.631E-04 |
|---|
| regulation of coagulation | GO:0050818 |  | 1.221E-06 | 5.875E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.032E-06 | 2.018E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 1.748E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.087E-06 | 1.53E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 5.087E-06 | 1.36E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 1.224E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.087E-06 | 1.113E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 5.775E-07 | 1.328E-03 |
|---|
| regulation of blood coagulation | GO:0030193 |  | 1.945E-06 | 2.236E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.49E-06 | 1.909E-03 |
|---|
| interphase | GO:0051325 |  | 2.731E-06 | 1.57E-03 |
|---|
| regulation of coagulation | GO:0050818 |  | 3.274E-06 | 1.506E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.241E-05 | 4.757E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.241E-05 | 4.077E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 1.241E-05 | 3.568E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.241E-05 | 3.171E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.535E-05 | 3.53E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 3.684E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 5.775E-07 | 1.328E-03 |
|---|
| regulation of blood coagulation | GO:0030193 |  | 1.945E-06 | 2.236E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.49E-06 | 1.909E-03 |
|---|
| interphase | GO:0051325 |  | 2.731E-06 | 1.57E-03 |
|---|
| regulation of coagulation | GO:0050818 |  | 3.274E-06 | 1.506E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.241E-05 | 4.757E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.241E-05 | 4.077E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 1.241E-05 | 3.568E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.241E-05 | 3.171E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.535E-05 | 3.53E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 3.684E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of blood coagulation | GO:0030193 |  | 5.595E-07 | 1.031E-03 |
|---|
| regulation of coagulation | GO:0050818 |  | 8.597E-07 | 7.922E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 6.833E-06 | 4.198E-03 |
|---|
| zymogen activation | GO:0031638 |  | 1.518E-05 | 6.995E-03 |
|---|
| regulation of fibrinolysis | GO:0051917 |  | 1.518E-05 | 5.596E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.518E-05 | 4.663E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.276E-05 | 5.992E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.276E-05 | 5.243E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 2.276E-05 | 4.66E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 2.276E-05 | 4.194E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 2.943E-05 | 4.93E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skin development | GO:0043588 |  | 5.775E-07 | 1.328E-03 |
|---|
| regulation of blood coagulation | GO:0030193 |  | 1.945E-06 | 2.236E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.49E-06 | 1.909E-03 |
|---|
| interphase | GO:0051325 |  | 2.731E-06 | 1.57E-03 |
|---|
| regulation of coagulation | GO:0050818 |  | 3.274E-06 | 1.506E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.241E-05 | 4.757E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.241E-05 | 4.077E-03 |
|---|
| plasminogen activation | GO:0031639 |  | 1.241E-05 | 3.568E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 1.241E-05 | 3.171E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.535E-05 | 3.53E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 3.684E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 1.752E-08 | 4.157E-05 |
|---|
| regulation of blood coagulation | GO:0030193 |  | 2.028E-08 | 2.406E-05 |
|---|
| regulation of coagulation | GO:0050818 |  | 3.895E-08 | 3.081E-05 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 6.121E-08 | 3.631E-05 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 1.467E-07 | 6.96E-05 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 1.467E-07 | 5.8E-05 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 2.093E-07 | 7.096E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.093E-07 | 6.209E-05 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 3.831E-07 | 1.01E-04 |
|---|
| negative regulation of lipid metabolic process | GO:0045833 |  | 4.977E-07 | 1.181E-04 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 4.977E-07 | 1.074E-04 |
|---|



