Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 22907-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1782 | 9.116e-03 | 6.743e-03 | 8.098e-02 |
|---|
| IPC-NIBC-129 | 0.2490 | 6.793e-02 | 9.634e-02 | 5.342e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2543 | 6.132e-03 | 1.064e-02 | 2.065e-01 |
|---|
| Loi_GPL570 | 0.1576 | 2.071e-01 | 2.258e-01 | 5.940e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1560 | 3.000e-06 | 3.200e-05 | 9.000e-06 |
|---|
| Parker_GPL1390 | 0.2623 | 4.890e-02 | 6.082e-02 | 3.892e-01 |
|---|
| Parker_GPL887 | 0.1776 | 4.106e-01 | 4.297e-01 | 4.548e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3210 | 2.589e-01 | 1.935e-01 | 8.279e-01 |
|---|
| Schmidt | 0.2445 | 2.889e-02 | 3.422e-02 | 2.981e-01 |
|---|
| Sotiriou | 0.3136 | 1.186e-02 | 9.115e-03 | 2.075e-01 |
|---|
| Van-De-Vijver | 0.2344 | 1.374e-03 | 1.977e-03 | 7.247e-02 |
|---|
| Zhang | 0.2483 | 1.427e-01 | 9.506e-02 | 2.952e-01 |
|---|
| Zhou | 0.4026 | 9.470e-02 | 8.660e-02 | 5.011e-01 |
|---|
Expression data for subnetwork 22907-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
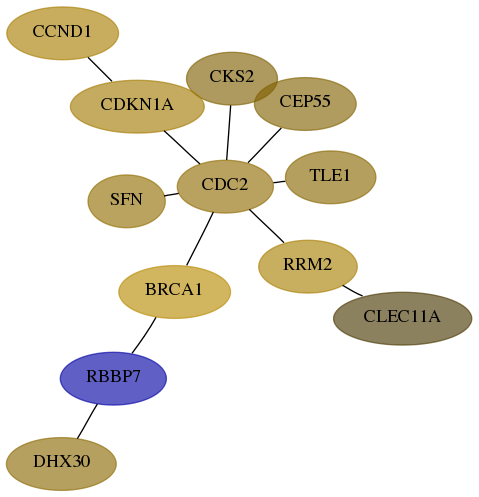
Score for each gene in subnetwork 22907-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| rrm2 |   | 5 | 40 | 163 | 143 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | undef | 0.129 | 0.302 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| cep55 |   | 6 | 35 | 207 | 187 | 0.054 | 0.210 | 0.148 | 0.129 | 0.135 | 0.162 | 0.133 | 0.209 | 0.202 | 0.138 | 0.189 | 0.115 | 0.330 |
|---|
| brca1 |   | 2 | 78 | 238 | 230 | 0.192 | 0.052 | 0.112 | 0.022 | 0.125 | -0.124 | 0.305 | 0.113 | 0.077 | 0.168 | 0.031 | 0.064 | 0.209 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| cks2 |   | 12 | 18 | 34 | 32 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.080 | 0.284 | 0.383 |
|---|
| rbbp7 |   | 1 | 116 | 238 | 240 | -0.113 | 0.089 | 0.051 | 0.076 | 0.106 | -0.053 | 0.092 | -0.020 | 0.126 | 0.138 | 0.069 | 0.168 | 0.280 |
|---|
| dhx30 |   | 2 | 78 | 120 | 117 | 0.066 | -0.039 | 0.122 | -0.072 | 0.099 | 0.272 | -0.261 | 0.326 | 0.065 | 0.160 | 0.026 | 0.015 | 0.304 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| clec11a |   | 2 | 78 | 163 | 156 | 0.013 | 0.101 | 0.071 | -0.085 | 0.066 | 0.082 | 0.209 | -0.126 | -0.034 | -0.001 | -0.039 | 0.059 | -0.075 |
|---|
GO Enrichment output for subnetwork 22907-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.587E-09 | 1.285E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.968E-08 | 4.563E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.938E-08 | 5.319E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.016E-07 | 1.159E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.605E-06 | 1.198E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.463E-06 | 1.327E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.652E-05 | 5.427E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.372E-05 | 6.819E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.476E-05 | 6.327E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 2.476E-05 | 5.694E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 2.476E-05 | 5.176E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.593E-10 | 1.122E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.78E-08 | 3.396E-05 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 6.997E-07 | 5.698E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.573E-06 | 2.793E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 4.743E-06 | 2.318E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 7.112E-06 | 2.896E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 7.112E-06 | 2.482E-03 |
|---|
| response to UV | GO:0009411 |  | 7.871E-06 | 2.404E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 9.952E-06 | 2.701E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 9.952E-06 | 2.431E-03 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.131E-05 | 2.512E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.295E-09 | 3.115E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 2.399E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 3.834E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.345E-08 | 3.817E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.7E-07 | 4.187E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.297E-06 | 5.202E-04 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 7.155E-06 | 2.459E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.459E-06 | 2.544E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.073E-05 | 2.868E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 1.073E-05 | 2.581E-03 |
|---|
| response to UV | GO:0009411 |  | 1.3E-05 | 2.844E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.593E-10 | 1.122E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.78E-08 | 3.396E-05 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 6.997E-07 | 5.698E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.573E-06 | 2.793E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 4.743E-06 | 2.318E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 7.112E-06 | 2.896E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 7.112E-06 | 2.482E-03 |
|---|
| response to UV | GO:0009411 |  | 7.871E-06 | 2.404E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 9.952E-06 | 2.701E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 9.952E-06 | 2.431E-03 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.131E-05 | 2.512E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.295E-09 | 3.115E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 2.399E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 3.834E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.345E-08 | 3.817E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.7E-07 | 4.187E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.297E-06 | 5.202E-04 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 7.155E-06 | 2.459E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.459E-06 | 2.544E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.073E-05 | 2.868E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 1.073E-05 | 2.581E-03 |
|---|
| response to UV | GO:0009411 |  | 1.3E-05 | 2.844E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.533E-11 | 1.976E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.667E-10 | 2.977E-07 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 5.971E-10 | 7.107E-07 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.54E-09 | 1.375E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.73E-08 | 1.236E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.044E-08 | 1.217E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.122E-07 | 5.723E-05 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 6.212E-07 | 2.773E-04 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 1.004E-06 | 3.983E-04 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 1.243E-06 | 4.439E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.618E-06 | 1.499E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.163E-09 | 4.242E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.416E-09 | 4.407E-06 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 9.093E-09 | 1.106E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.866E-08 | 1.702E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 6.789E-08 | 4.953E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.153E-07 | 7.013E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 6.749E-07 | 3.517E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.848E-06 | 8.427E-04 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 7.119E-06 | 2.886E-03 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 7.781E-06 | 2.839E-03 |
|---|
| negative regulation of fatty acid metabolic process | GO:0045922 |  | 1.196E-05 | 3.967E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.295E-09 | 3.115E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 2.399E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 3.834E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.345E-08 | 3.817E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.7E-07 | 4.187E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.297E-06 | 5.202E-04 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 7.155E-06 | 2.459E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.459E-06 | 2.544E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.073E-05 | 2.868E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 1.073E-05 | 2.581E-03 |
|---|
| response to UV | GO:0009411 |  | 1.3E-05 | 2.844E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.587E-09 | 1.285E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.968E-08 | 4.563E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.938E-08 | 5.319E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.016E-07 | 1.159E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.605E-06 | 1.198E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.463E-06 | 1.327E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.652E-05 | 5.427E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.372E-05 | 6.819E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.476E-05 | 6.327E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 2.476E-05 | 5.694E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 2.476E-05 | 5.176E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.587E-09 | 1.285E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.968E-08 | 4.563E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.938E-08 | 5.319E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.016E-07 | 1.159E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.605E-06 | 1.198E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.463E-06 | 1.327E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.652E-05 | 5.427E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.372E-05 | 6.819E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.476E-05 | 6.327E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 2.476E-05 | 5.694E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 2.476E-05 | 5.176E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.251E-06 | 2.306E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.965E-06 | 3.654E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.923E-06 | 6.096E-03 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.177E-05 | 5.421E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 1.518E-05 | 5.596E-03 |
|---|
| postreplication repair | GO:0006301 |  | 1.518E-05 | 4.663E-03 |
|---|
| regulation of transcription from RNA polymerase III promoter | GO:0006359 |  | 1.518E-05 | 3.997E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.276E-05 | 5.243E-03 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 3.184E-05 | 6.52E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.184E-05 | 5.868E-03 |
|---|
| negative regulation of lipid metabolic process | GO:0045833 |  | 3.184E-05 | 5.335E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.587E-09 | 1.285E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.968E-08 | 4.563E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.938E-08 | 5.319E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.016E-07 | 1.159E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.605E-06 | 1.198E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.463E-06 | 1.327E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.652E-05 | 5.427E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.372E-05 | 6.819E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.476E-05 | 6.327E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 2.476E-05 | 5.694E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 2.476E-05 | 5.176E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.651E-09 | 1.341E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 6.121E-08 | 7.262E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 9.785E-08 | 7.74E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.104E-07 | 1.248E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.66E-06 | 1.262E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.49E-06 | 1.38E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 1.514E-05 | 5.131E-03 |
|---|
| regulation of centrosome duplication | GO:0010824 |  | 1.514E-05 | 4.489E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.246E-05 | 5.922E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 2.269E-05 | 5.383E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.269E-05 | 4.894E-03 |
|---|



