Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1501-1-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1723 | 1.804e-02 | 1.461e-02 | 1.381e-01 |
|---|
| IPC-NIBC-129 | 0.2323 | 4.310e-02 | 6.244e-02 | 4.046e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2211 | 7.035e-03 | 1.207e-02 | 2.236e-01 |
|---|
| Loi_GPL570 | 0.2503 | 5.833e-02 | 6.610e-02 | 2.467e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1586 | 1.000e-05 | 8.900e-05 | 6.300e-05 |
|---|
| Parker_GPL1390 | 0.2560 | 3.007e-02 | 3.824e-02 | 2.948e-01 |
|---|
| Parker_GPL887 | 0.2396 | 4.253e-01 | 4.444e-01 | 4.728e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.2878 | 1.354e-01 | 8.928e-02 | 6.307e-01 |
|---|
| Schmidt | 0.2783 | 8.607e-03 | 9.360e-03 | 1.282e-01 |
|---|
| Sotiriou | 0.3953 | 2.997e-03 | 2.217e-03 | 9.171e-02 |
|---|
| Van-De-Vijver | 0.2678 | 9.520e-04 | 1.369e-03 | 5.677e-02 |
|---|
| Zhang | 0.3217 | 1.370e-01 | 9.036e-02 | 2.856e-01 |
|---|
| Zhou | 0.4231 | 1.275e-01 | 1.262e-01 | 6.019e-01 |
|---|
Expression data for subnetwork 1501-1-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
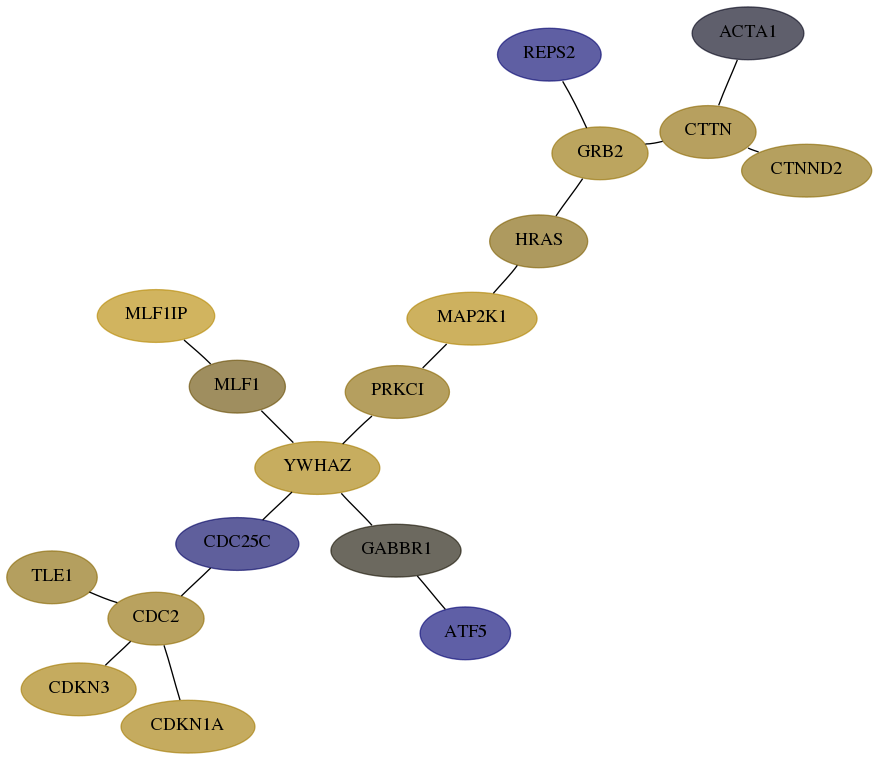
Score for each gene in subnetwork 1501-1-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| cttn |   | 10 | 20 | 1 | 6 | 0.067 | 0.065 | 0.123 | 0.238 | 0.126 | 0.263 | -0.172 | 0.216 | 0.069 | 0.115 | undef | 0.025 | 0.279 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| mlf1ip |   | 5 | 40 | 77 | 64 | 0.182 | 0.147 | 0.186 | 0.144 | 0.242 | 0.266 | 0.005 | 0.345 | 0.182 | 0.197 | undef | 0.240 | 0.372 |
|---|
| prkci |   | 14 | 15 | 31 | 26 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | 0.049 | 0.124 | 0.149 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| hras |   | 14 | 15 | 42 | 34 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.195 | 0.022 | 0.245 | 0.086 | 0.208 | 0.180 | 0.003 | 0.197 |
|---|
| gabbr1 |   | 7 | 30 | 56 | 49 | 0.003 | -0.016 | -0.022 | -0.051 | 0.082 | 0.119 | 0.584 | -0.082 | 0.049 | 0.057 | 0.125 | 0.096 | 0.116 |
|---|
| ctnnd2 |   | 1 | 116 | 81 | 91 | 0.065 | -0.042 | -0.017 | 0.120 | 0.136 | -0.062 | 0.238 | -0.047 | -0.082 | 0.133 | undef | 0.141 | -0.079 |
|---|
| map2k1 |   | 10 | 20 | 54 | 44 | 0.154 | 0.096 | 0.093 | 0.013 | 0.095 | 0.045 | -0.223 | 0.207 | 0.082 | 0.096 | undef | undef | 0.230 |
|---|
| atf5 |   | 1 | 116 | 81 | 91 | -0.036 | 0.163 | 0.158 | 0.045 | 0.106 | 0.007 | 0.132 | 0.113 | 0.056 | 0.199 | undef | 0.048 | 0.202 |
|---|
| mlf1 |   | 7 | 30 | 56 | 49 | 0.027 | 0.224 | 0.053 | 0.047 | 0.169 | 0.128 | 0.021 | -0.028 | 0.106 | 0.176 | undef | 0.098 | 0.141 |
|---|
| reps2 |   | 3 | 62 | 56 | 56 | -0.033 | -0.139 | 0.057 | 0.122 | 0.113 | -0.154 | 0.105 | -0.157 | 0.079 | 0.124 | 0.052 | 0.303 | 0.220 |
|---|
| cdc25c |   | 2 | 78 | 81 | 80 | -0.025 | 0.187 | 0.084 | 0.267 | 0.070 | 0.059 | 0.367 | 0.125 | 0.204 | 0.250 | 0.086 | 0.054 | 0.351 |
|---|
| ywhaz |   | 17 | 10 | 29 | 25 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | undef | 0.103 | 0.213 |
|---|
| acta1 |   | 9 | 22 | 1 | 7 | -0.003 | 0.098 | -0.008 | -0.030 | 0.119 | 0.174 | 0.376 | 0.024 | 0.182 | 0.214 | 0.100 | 0.048 | 0.201 |
|---|
| grb2 |   | 9 | 22 | 1 | 7 | 0.084 | -0.013 | 0.086 | -0.031 | 0.162 | -0.038 | 0.003 | 0.065 | 0.158 | 0.127 | 0.149 | 0.194 | -0.005 |
|---|
GO Enrichment output for subnetwork 1501-1-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 6.703E-08 | 1.542E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.097E-06 | 1.262E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.793E-06 | 2.141E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.699E-06 | 2.127E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 7.491E-06 | 3.446E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.323E-05 | 5.071E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.695E-05 | 5.569E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.262E-05 | 6.502E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.262E-05 | 5.78E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.372E-05 | 5.457E-03 |
|---|
| interphase | GO:0051325 |  | 2.477E-05 | 5.179E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.646E-08 | 4.022E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.677E-07 | 5.713E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.998E-07 | 5.699E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 9.134E-07 | 5.579E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.16E-06 | 1.055E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 4.205E-06 | 1.712E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.205E-06 | 1.468E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 5.29E-06 | 1.615E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.29E-06 | 1.436E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.173E-06 | 1.508E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 7.982E-06 | 1.773E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.517E-08 | 6.056E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.142E-07 | 8.592E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.223E-06 | 9.807E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.394E-06 | 8.388E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.828E-06 | 1.361E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.001E-06 | 2.005E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.677E-06 | 1.951E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 5.677E-06 | 1.707E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 7.206E-06 | 1.926E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.485E-06 | 2.282E-03 |
|---|
| interphase | GO:0051325 |  | 1.073E-05 | 2.347E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.646E-08 | 4.022E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.677E-07 | 5.713E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.998E-07 | 5.699E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 9.134E-07 | 5.579E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.16E-06 | 1.055E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 4.205E-06 | 1.712E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.205E-06 | 1.468E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 5.29E-06 | 1.615E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.29E-06 | 1.436E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.173E-06 | 1.508E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 7.982E-06 | 1.773E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.517E-08 | 6.056E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.142E-07 | 8.592E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.223E-06 | 9.807E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.394E-06 | 8.388E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.828E-06 | 1.361E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.001E-06 | 2.005E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.677E-06 | 1.951E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 5.677E-06 | 1.707E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 7.206E-06 | 1.926E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.485E-06 | 2.282E-03 |
|---|
| interphase | GO:0051325 |  | 1.073E-05 | 2.347E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.467E-07 | 1.238E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.569E-06 | 4.588E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 3.258E-06 | 3.878E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 3.463E-06 | 3.091E-03 |
|---|
| protein tyrosine phosphatase activity | GO:0004725 |  | 4.074E-06 | 2.909E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 5.459E-06 | 3.249E-03 |
|---|
| regulation of synaptic plasticity | GO:0048167 |  | 5.484E-06 | 2.798E-03 |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 6.736E-06 | 3.007E-03 |
|---|
| learning | GO:0007612 |  | 1.257E-05 | 4.989E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 1.407E-05 | 5.023E-03 |
|---|
| insulin receptor substrate binding | GO:0043560 |  | 2.108E-05 | 6.844E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.292E-07 | 8.361E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.495E-06 | 2.727E-03 |
|---|
| interphase | GO:0051325 |  | 1.649E-06 | 2.005E-03 |
|---|
| protein tyrosine phosphatase activity | GO:0004725 |  | 2.182E-06 | 1.99E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.602E-06 | 1.898E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 2.602E-06 | 1.582E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 2.669E-06 | 1.391E-03 |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 3.744E-06 | 1.707E-03 |
|---|
| neuron adhesion | GO:0007158 |  | 1.082E-05 | 4.387E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 1.082E-05 | 3.948E-03 |
|---|
| muscle thin filament assembly | GO:0030240 |  | 1.082E-05 | 3.589E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.517E-08 | 6.056E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.142E-07 | 8.592E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.223E-06 | 9.807E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.394E-06 | 8.388E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.828E-06 | 1.361E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.001E-06 | 2.005E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.677E-06 | 1.951E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 5.677E-06 | 1.707E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 7.206E-06 | 1.926E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.485E-06 | 2.282E-03 |
|---|
| interphase | GO:0051325 |  | 1.073E-05 | 2.347E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 6.703E-08 | 1.542E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.097E-06 | 1.262E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.793E-06 | 2.141E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.699E-06 | 2.127E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 7.491E-06 | 3.446E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.323E-05 | 5.071E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.695E-05 | 5.569E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.262E-05 | 6.502E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.262E-05 | 5.78E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.372E-05 | 5.457E-03 |
|---|
| interphase | GO:0051325 |  | 2.477E-05 | 5.179E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 6.703E-08 | 1.542E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.097E-06 | 1.262E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.793E-06 | 2.141E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.699E-06 | 2.127E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 7.491E-06 | 3.446E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.323E-05 | 5.071E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.695E-05 | 5.569E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.262E-05 | 6.502E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.262E-05 | 5.78E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.372E-05 | 5.457E-03 |
|---|
| interphase | GO:0051325 |  | 2.477E-05 | 5.179E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 4.666E-07 | 8.599E-04 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 8.354E-07 | 7.698E-04 |
|---|
| muscle cell differentiation | GO:0042692 |  | 6.073E-06 | 3.731E-03 |
|---|
| regulation of long-term neuronal synaptic plasticity | GO:0048169 |  | 1.181E-05 | 5.442E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 1.181E-05 | 4.354E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 1.181E-05 | 3.628E-03 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.181E-05 | 3.11E-03 |
|---|
| actin filament organization | GO:0007015 |  | 1.742E-05 | 4.013E-03 |
|---|
| establishment or maintenance of apical/basal cell polarity | GO:0035088 |  | 1.771E-05 | 3.626E-03 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 1.771E-05 | 3.263E-03 |
|---|
| visual behavior | GO:0007632 |  | 2.478E-05 | 4.151E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 6.703E-08 | 1.542E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.097E-06 | 1.262E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.793E-06 | 2.141E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.699E-06 | 2.127E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 7.491E-06 | 3.446E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.323E-05 | 5.071E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.695E-05 | 5.569E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.262E-05 | 6.502E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.262E-05 | 5.78E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.372E-05 | 5.457E-03 |
|---|
| interphase | GO:0051325 |  | 2.477E-05 | 5.179E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.981E-08 | 1.182E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.087E-06 | 1.289E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.213E-06 | 1.751E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.338E-06 | 1.98E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 7.515E-06 | 3.566E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.263E-05 | 4.994E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.263E-05 | 4.28E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.613E-05 | 4.783E-03 |
|---|
| interphase | GO:0051325 |  | 1.764E-05 | 4.652E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.099E-05 | 4.982E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.17E-05 | 4.681E-03 |
|---|



