Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 9024-10-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2038 | 9.441e-03 | 1.128e-02 | 9.490e-02 |
|---|
| IPC-NIBC-129 | 0.2395 | 5.255e-02 | 6.711e-02 | 4.674e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2511 | 9.807e-03 | 1.513e-02 | 2.603e-01 |
|---|
| Loi_GPL570 | 0.2439 | 7.092e-02 | 7.045e-02 | 3.277e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1564 | 9.000e-06 | 5.900e-05 | 6.400e-05 |
|---|
| Parker_GPL1390 | 0.2547 | 9.319e-02 | 9.232e-02 | 5.226e-01 |
|---|
| Parker_GPL887 | 0.0579 | 8.096e-01 | 8.278e-01 | 8.631e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3107 | 1.372e-01 | 1.055e-01 | 6.206e-01 |
|---|
| Schmidt | 0.2408 | 9.785e-03 | 1.399e-02 | 1.286e-01 |
|---|
| Sotiriou | 0.3165 | 1.440e-03 | 3.600e-04 | 4.158e-02 |
|---|
| Wang | 0.2155 | 1.173e-01 | 8.286e-02 | 4.051e-01 |
|---|
| Zhang | 0.2402 | 6.276e-02 | 3.749e-02 | 1.574e-01 |
|---|
| Zhou | 0.4460 | 9.763e-02 | 1.076e-01 | 5.426e-01 |
|---|
Expression data for subnetwork 9024-10-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
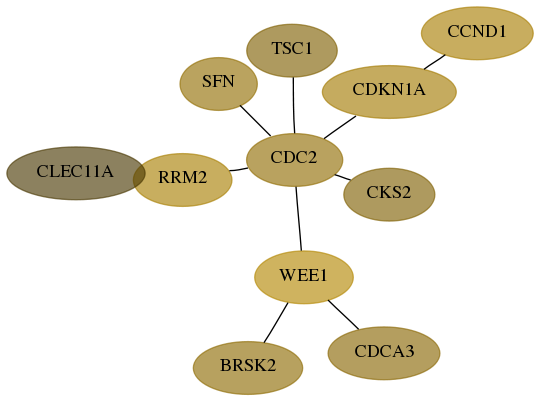
Score for each gene in subnetwork 9024-10-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| rrm2 |   | 21 | 10 | 44 | 26 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | 0.117 | 0.129 | 0.302 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| brsk2 |   | 1 | 164 | 289 | 294 | 0.068 | 0.122 | 0.012 | 0.075 | 0.079 | 0.056 | 0.128 | -0.069 | 0.068 | 0.096 | -0.023 | -0.042 | 0.022 |
|---|
| wee1 |   | 2 | 104 | 289 | 284 | 0.169 | -0.036 | -0.023 | 0.062 | 0.083 | 0.129 | -0.162 | 0.112 | 0.043 | 0.033 | 0.086 | 0.008 | 0.328 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
| clec11a |   | 17 | 12 | 44 | 27 | 0.013 | 0.101 | 0.071 | -0.085 | 0.066 | 0.082 | 0.209 | -0.126 | -0.034 | -0.001 | 0.191 | 0.059 | -0.075 |
|---|
| cdca3 |   | 2 | 104 | 289 | 284 | 0.061 | 0.245 | 0.116 | 0.187 | 0.152 | -0.141 | 0.224 | 0.247 | 0.234 | 0.117 | 0.086 | 0.102 | 0.401 |
|---|
GO Enrichment output for subnetwork 9024-10-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.041E-09 | 2.394E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.525E-08 | 1.754E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 2.667E-08 | 2.045E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.005E-06 | 5.778E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.38E-06 | 3.395E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.885E-06 | 3.406E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 8.885E-06 | 2.919E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 2.646E-03 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 2.887E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.287E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.864E-05 | 3.897E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.471E-11 | 1.825E-07 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.527E-06 | 1.865E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.633E-06 | 1.33E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.42E-06 | 1.478E-03 |
|---|
| response to UV | GO:0009411 |  | 2.814E-06 | 1.375E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.629E-06 | 1.478E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.079E-06 | 1.773E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 5.595E-06 | 1.709E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 5.843E-06 | 1.586E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.77E-06 | 1.654E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.77E-06 | 1.504E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.312E-10 | 5.562E-07 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 7.438E-09 | 8.948E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.784E-08 | 1.431E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.253E-07 | 1.957E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.97E-06 | 1.429E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.176E-06 | 1.274E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.766E-06 | 1.295E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.766E-06 | 1.133E-03 |
|---|
| response to UV | GO:0009411 |  | 4.888E-06 | 1.307E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 9.94E-06 | 2.392E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.039E-05 | 2.274E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.471E-11 | 1.825E-07 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.527E-06 | 1.865E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.633E-06 | 1.33E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.42E-06 | 1.478E-03 |
|---|
| response to UV | GO:0009411 |  | 2.814E-06 | 1.375E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.629E-06 | 1.478E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.079E-06 | 1.773E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 5.595E-06 | 1.709E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 5.843E-06 | 1.586E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.77E-06 | 1.654E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.77E-06 | 1.504E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.312E-10 | 5.562E-07 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 7.438E-09 | 8.948E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.784E-08 | 1.431E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.253E-07 | 1.957E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.97E-06 | 1.429E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.176E-06 | 1.274E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.766E-06 | 1.295E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.766E-06 | 1.133E-03 |
|---|
| response to UV | GO:0009411 |  | 4.888E-06 | 1.307E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 9.94E-06 | 2.392E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.039E-05 | 2.274E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.522E-11 | 1.258E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.116E-11 | 1.628E-07 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 3.268E-10 | 3.889E-07 |
|---|
| kinase regulator activity | GO:0019207 |  | 8.436E-10 | 7.531E-07 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.259E-08 | 8.989E-06 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 8.163E-08 | 4.858E-05 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 7.31E-07 | 3.729E-04 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 9.053E-07 | 4.041E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.739E-06 | 1.087E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.366E-06 | 1.202E-03 |
|---|
| response to UV | GO:0009411 |  | 4.081E-06 | 1.325E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.599E-11 | 2.042E-07 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 6.438E-11 | 1.174E-07 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 2.121E-10 | 2.58E-07 |
|---|
| kinase regulator activity | GO:0019207 |  | 4.374E-10 | 3.989E-07 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 8.439E-09 | 6.157E-06 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 8.417E-08 | 5.118E-05 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 8.943E-07 | 4.66E-04 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 9.778E-07 | 4.459E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.589E-06 | 6.443E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.839E-06 | 6.708E-04 |
|---|
| response to UV | GO:0009411 |  | 2.113E-06 | 7.008E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.312E-10 | 5.562E-07 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 7.438E-09 | 8.948E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.784E-08 | 1.431E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.253E-07 | 1.957E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.97E-06 | 1.429E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.176E-06 | 1.274E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.766E-06 | 1.295E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.766E-06 | 1.133E-03 |
|---|
| response to UV | GO:0009411 |  | 4.888E-06 | 1.307E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 9.94E-06 | 2.392E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.039E-05 | 2.274E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.041E-09 | 2.394E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.525E-08 | 1.754E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 2.667E-08 | 2.045E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.005E-06 | 5.778E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.38E-06 | 3.395E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.885E-06 | 3.406E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 8.885E-06 | 2.919E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 2.646E-03 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 2.887E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.287E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.864E-05 | 3.897E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.041E-09 | 2.394E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.525E-08 | 1.754E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 2.667E-08 | 2.045E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.005E-06 | 5.778E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.38E-06 | 3.395E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.885E-06 | 3.406E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 8.885E-06 | 2.919E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 2.646E-03 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 2.887E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.287E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.864E-05 | 3.897E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.041E-09 | 2.394E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.525E-08 | 1.754E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 2.667E-08 | 2.045E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.005E-06 | 5.778E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.38E-06 | 3.395E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.885E-06 | 3.406E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 8.885E-06 | 2.919E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 2.646E-03 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 2.887E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.287E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.864E-05 | 3.897E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.041E-09 | 2.394E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.525E-08 | 1.754E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 2.667E-08 | 2.045E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.005E-06 | 5.778E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.38E-06 | 3.395E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.885E-06 | 3.406E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 8.885E-06 | 2.919E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 2.646E-03 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 2.887E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.287E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.864E-05 | 3.897E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.666E-09 | 3.953E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.036E-08 | 3.602E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.854E-08 | 3.84E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.323E-06 | 7.849E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 9.59E-06 | 4.552E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.737E-06 | 3.851E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.122E-05 | 3.803E-03 |
|---|
| response to UV | GO:0009411 |  | 1.371E-05 | 4.066E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.438E-05 | 3.791E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.012E-05 | 4.773E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.68E-05 | 5.782E-03 |
|---|



