Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 7415-9-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1713 | 8.734e-03 | 1.043e-02 | 8.950e-02 |
|---|
| IPC-NIBC-129 | 0.2815 | 7.520e-02 | 9.792e-02 | 5.806e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2548 | 2.347e-02 | 3.316e-02 | 4.228e-01 |
|---|
| Loi_GPL570 | 0.2028 | 3.986e-02 | 3.955e-02 | 2.151e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1646 | 1.100e-05 | 6.800e-05 | 8.400e-05 |
|---|
| Parker_GPL1390 | 0.2284 | 1.460e-01 | 1.501e-01 | 6.427e-01 |
|---|
| Parker_GPL887 | 0.1699 | 4.299e-01 | 4.646e-01 | 4.546e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3521 | 2.254e-01 | 1.874e-01 | 7.825e-01 |
|---|
| Schmidt | 0.2383 | 1.283e-02 | 1.813e-02 | 1.581e-01 |
|---|
| Sotiriou | 0.3000 | 4.353e-03 | 1.456e-03 | 9.244e-02 |
|---|
| Wang | 0.1891 | 1.028e-01 | 7.105e-02 | 3.722e-01 |
|---|
| Zhang | 0.1903 | 9.191e-02 | 5.921e-02 | 2.230e-01 |
|---|
| Zhou | 0.3794 | 6.611e-02 | 7.042e-02 | 4.036e-01 |
|---|
Expression data for subnetwork 7415-9-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
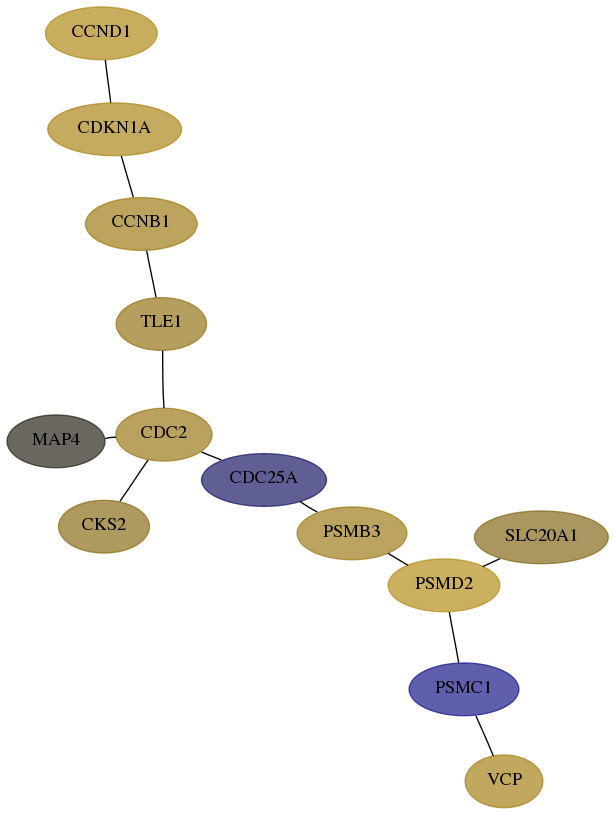
Score for each gene in subnetwork 7415-9-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| psmd2 |   | 7 | 38 | 37 | 28 | 0.147 | 0.322 | 0.122 | 0.146 | 0.143 | 0.246 | 0.444 | 0.194 | 0.318 | 0.151 | 0.154 | 0.042 | 0.207 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| tle1 |   | 23 | 9 | 78 | 60 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | -0.028 | 0.068 | 0.025 |
|---|
| vcp |   | 1 | 164 | 349 | 349 | 0.100 | 0.137 | 0.099 | 0.055 | 0.092 | 0.165 | 0.136 | 0.261 | -0.063 | 0.060 | -0.062 | -0.096 | 0.158 |
|---|
| ccnb1 |   | 36 | 7 | 78 | 59 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 | 0.398 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| psmc1 |   | 2 | 104 | 61 | 69 | -0.048 | 0.159 | 0.166 | 0.140 | 0.109 | 0.032 | 0.072 | 0.250 | 0.161 | 0.120 | 0.086 | 0.214 | 0.275 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| map4 |   | 16 | 13 | 73 | 55 | 0.002 | 0.046 | 0.067 | -0.043 | 0.034 | 0.119 | 0.159 | -0.058 | -0.020 | 0.043 | 0.108 | -0.074 | 0.302 |
|---|
| cdc25a |   | 9 | 27 | 109 | 90 | -0.019 | 0.221 | 0.093 | 0.158 | 0.120 | -0.061 | 0.406 | 0.207 | 0.211 | 0.016 | 0.069 | 0.183 | 0.264 |
|---|
| psmb3 |   | 5 | 50 | 150 | 138 | 0.080 | 0.214 | 0.172 | 0.037 | 0.177 | -0.031 | -0.058 | 0.133 | 0.154 | 0.132 | 0.038 | 0.047 | 0.059 |
|---|
| slc20a1 |   | 7 | 38 | 37 | 28 | 0.041 | 0.039 | 0.109 | 0.170 | 0.101 | 0.121 | 0.326 | 0.086 | 0.048 | 0.124 | 0.121 | -0.050 | 0.299 |
|---|
GO Enrichment output for subnetwork 7415-9-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.971E-11 | 4.533E-08 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.057E-10 | 3.516E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.382E-10 | 2.593E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.53E-10 | 2.605E-07 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.53E-10 | 2.084E-07 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 6.549E-10 | 2.51E-07 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 7.809E-10 | 2.566E-07 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.094E-09 | 3.144E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.085E-08 | 2.773E-06 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.615E-06 | 3.713E-04 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 1.771E-06 | 3.703E-04 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 2.695E-12 | 6.584E-09 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.159E-11 | 5.08E-08 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 4.552E-11 | 3.707E-08 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 5.921E-11 | 3.616E-08 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 5.921E-11 | 2.893E-08 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 8.956E-11 | 3.647E-08 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.049E-10 | 3.66E-08 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.528E-10 | 4.668E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.032E-09 | 5.516E-07 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 4.527E-07 | 1.106E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 4.807E-07 | 1.067E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 6.722E-12 | 1.617E-08 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 9.145E-11 | 1.1E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.001E-10 | 8.027E-08 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.302E-10 | 7.829E-08 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.302E-10 | 6.263E-08 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.968E-10 | 7.891E-08 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.304E-10 | 7.918E-08 |
|---|
| regulation of ligase activity | GO:0051340 |  | 3.356E-10 | 1.009E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.929E-09 | 1.05E-06 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 7.674E-07 | 1.846E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 8.147E-07 | 1.782E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 2.695E-12 | 6.584E-09 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.159E-11 | 5.08E-08 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 4.552E-11 | 3.707E-08 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 5.921E-11 | 3.616E-08 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 5.921E-11 | 2.893E-08 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 8.956E-11 | 3.647E-08 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.049E-10 | 3.66E-08 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.528E-10 | 4.668E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.032E-09 | 5.516E-07 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 4.527E-07 | 1.106E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 4.807E-07 | 1.067E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 6.722E-12 | 1.617E-08 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 9.145E-11 | 1.1E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.001E-10 | 8.027E-08 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.302E-10 | 7.829E-08 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.302E-10 | 6.263E-08 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.968E-10 | 7.891E-08 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.304E-10 | 7.918E-08 |
|---|
| regulation of ligase activity | GO:0051340 |  | 3.356E-10 | 1.009E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.929E-09 | 1.05E-06 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 7.674E-07 | 1.846E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 8.147E-07 | 1.782E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 2.575E-11 | 9.194E-08 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 8.297E-11 | 1.481E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.851E-10 | 3.393E-07 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 7.969E-10 | 7.114E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.02E-09 | 7.286E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.105E-09 | 6.575E-07 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.291E-09 | 6.584E-07 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.501E-09 | 6.699E-07 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.866E-09 | 7.404E-07 |
|---|
| regulation of ligase activity | GO:0051340 |  | 2.46E-09 | 8.785E-07 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.306E-08 | 7.487E-06 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 4.368E-14 | 1.593E-10 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.735E-12 | 3.164E-09 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.329E-12 | 2.832E-09 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.812E-12 | 2.564E-09 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 3.083E-12 | 2.249E-09 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 3.691E-12 | 2.244E-09 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 5.659E-12 | 2.949E-09 |
|---|
| regulation of ligase activity | GO:0051340 |  | 7.211E-12 | 3.288E-09 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.394E-10 | 5.649E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.553E-10 | 5.666E-08 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.462E-08 | 4.849E-06 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 6.722E-12 | 1.617E-08 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 9.145E-11 | 1.1E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.001E-10 | 8.027E-08 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.302E-10 | 7.829E-08 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.302E-10 | 6.263E-08 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.968E-10 | 7.891E-08 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.304E-10 | 7.918E-08 |
|---|
| regulation of ligase activity | GO:0051340 |  | 3.356E-10 | 1.009E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.929E-09 | 1.05E-06 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 7.674E-07 | 1.846E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 8.147E-07 | 1.782E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.971E-11 | 4.533E-08 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.057E-10 | 3.516E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.382E-10 | 2.593E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.53E-10 | 2.605E-07 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.53E-10 | 2.084E-07 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 6.549E-10 | 2.51E-07 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 7.809E-10 | 2.566E-07 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.094E-09 | 3.144E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.085E-08 | 2.773E-06 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.615E-06 | 3.713E-04 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 1.771E-06 | 3.703E-04 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.971E-11 | 4.533E-08 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.057E-10 | 3.516E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.382E-10 | 2.593E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.53E-10 | 2.605E-07 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.53E-10 | 2.084E-07 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 6.549E-10 | 2.51E-07 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 7.809E-10 | 2.566E-07 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.094E-09 | 3.144E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.085E-08 | 2.773E-06 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.615E-06 | 3.713E-04 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 1.771E-06 | 3.703E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.971E-11 | 4.533E-08 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.057E-10 | 3.516E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.382E-10 | 2.593E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.53E-10 | 2.605E-07 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.53E-10 | 2.084E-07 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 6.549E-10 | 2.51E-07 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 7.809E-10 | 2.566E-07 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.094E-09 | 3.144E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.085E-08 | 2.773E-06 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.615E-06 | 3.713E-04 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 1.771E-06 | 3.703E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.971E-11 | 4.533E-08 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.057E-10 | 3.516E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.382E-10 | 2.593E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.53E-10 | 2.605E-07 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.53E-10 | 2.084E-07 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 6.549E-10 | 2.51E-07 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 7.809E-10 | 2.566E-07 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.094E-09 | 3.144E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.085E-08 | 2.773E-06 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.615E-06 | 3.713E-04 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 1.771E-06 | 3.703E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.247E-11 | 2.959E-08 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.313E-10 | 2.744E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.326E-10 | 2.631E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.307E-10 | 2.555E-07 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.307E-10 | 2.044E-07 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 5.517E-10 | 2.182E-07 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 6.997E-10 | 2.372E-07 |
|---|
| regulation of ligase activity | GO:0051340 |  | 8.791E-10 | 2.608E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.982E-09 | 1.841E-06 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 1.565E-06 | 3.714E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.598E-06 | 3.446E-04 |
|---|



