Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6929-8-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2523 | 7.459e-03 | 8.897e-03 | 7.939e-02 |
|---|
| IPC-NIBC-129 | 0.2516 | 5.166e-02 | 6.591e-02 | 4.622e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2625 | 6.703e-03 | 1.074e-02 | 2.058e-01 |
|---|
| Loi_GPL570 | 0.2617 | 6.012e-02 | 5.970e-02 | 2.915e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1681 | 1.600e-05 | 9.700e-05 | 1.510e-04 |
|---|
| Parker_GPL1390 | 0.2559 | 4.699e-02 | 4.362e-02 | 3.640e-01 |
|---|
| Parker_GPL887 | 0.1555 | 4.695e-01 | 5.038e-01 | 5.004e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3702 | 4.315e-01 | 3.953e-01 | 9.457e-01 |
|---|
| Schmidt | 0.2327 | 1.610e-02 | 2.252e-02 | 1.872e-01 |
|---|
| Sotiriou | 0.2860 | 1.135e-03 | 2.660e-04 | 3.479e-02 |
|---|
| Wang | 0.2173 | 9.727e-02 | 6.658e-02 | 3.589e-01 |
|---|
| Zhang | 0.1214 | 6.109e-02 | 3.630e-02 | 1.535e-01 |
|---|
| Zhou | 0.4602 | 5.561e-02 | 5.826e-02 | 3.485e-01 |
|---|
Expression data for subnetwork 6929-8-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
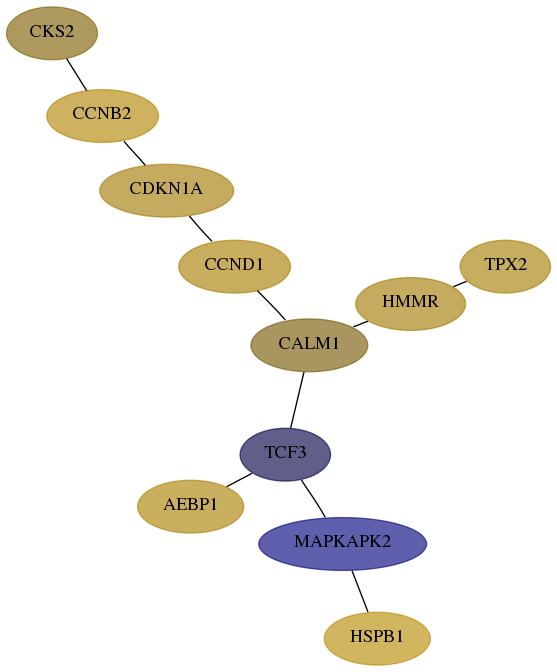
Score for each gene in subnetwork 6929-8-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| ccnb2 |   | 16 | 13 | 153 | 125 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.178 | 0.090 | 0.366 | 0.240 | 0.169 | 0.144 | 0.211 | 0.463 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| tpx2 |   | 2 | 104 | 166 | 158 | 0.125 | 0.234 | 0.143 | 0.235 | 0.185 | 0.062 | 0.375 | 0.317 | 0.210 | 0.209 | 0.028 | 0.103 | 0.411 |
|---|
| tcf3 |   | 3 | 79 | 17 | 22 | -0.012 | 0.282 | 0.055 | 0.045 | 0.037 | 0.176 | 0.318 | 0.006 | 0.094 | -0.056 | 0.040 | 0.001 | 0.167 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| calm1 |   | 4 | 60 | 130 | 115 | 0.038 | 0.023 | 0.171 | 0.277 | 0.113 | 0.034 | -0.199 | 0.007 | 0.171 | 0.112 | 0.087 | 0.162 | 0.171 |
|---|
| hmmr |   | 4 | 60 | 130 | 115 | 0.112 | 0.310 | 0.147 | 0.127 | 0.145 | 0.134 | 0.131 | 0.221 | 0.179 | 0.233 | 0.227 | 0.144 | 0.325 |
|---|
| aebp1 |   | 1 | 164 | 166 | 177 | 0.139 | 0.166 | 0.024 | 0.007 | 0.047 | 0.144 | -0.178 | -0.029 | -0.086 | -0.032 | 0.130 | -0.020 | 0.160 |
|---|
| mapkapk2 |   | 13 | 19 | 17 | 12 | -0.050 | 0.102 | 0.156 | -0.116 | 0.090 | 0.168 | 0.303 | 0.074 | 0.171 | 0.117 | 0.085 | -0.124 | 0.312 |
|---|
GO Enrichment output for subnetwork 6929-8-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.018E-07 | 2.342E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.159E-07 | 1.053E-03 |
|---|
| B cell differentiation | GO:0030183 |  | 2.782E-05 | 0.02132696 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.464E-05 | 0.0199204 |
|---|
| response to UV | GO:0009411 |  | 4.249E-05 | 0.01954448 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01704369 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 5.923E-05 | 0.01946068 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 5.923E-05 | 0.01702809 |
|---|
| thymus development | GO:0048538 |  | 7.608E-05 | 0.01944287 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 9.501E-05 | 0.02185322 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 1.146E-04 | 0.02395412 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 2.097E-08 | 5.124E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.81E-07 | 2.211E-04 |
|---|
| B cell differentiation | GO:0030183 |  | 8.314E-06 | 6.771E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.889E-06 | 5.429E-03 |
|---|
| response to UV | GO:0009411 |  | 1.528E-05 | 7.466E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.054E-05 | 8.365E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.298E-05 | 0.01151093 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.298E-05 | 0.01007206 |
|---|
| B cell activation | GO:0042113 |  | 3.439E-05 | 9.335E-03 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 3.734E-05 | 9.121E-03 |
|---|
| thymus development | GO:0048538 |  | 4.029E-05 | 8.948E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 2.512E-08 | 6.045E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.281E-07 | 2.745E-04 |
|---|
| B cell differentiation | GO:0030183 |  | 9.29E-06 | 7.451E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.064E-05 | 6.397E-03 |
|---|
| response to UV | GO:0009411 |  | 1.634E-05 | 7.863E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.326E-05 | 9.326E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.734E-05 | 0.01283284 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.734E-05 | 0.01122874 |
|---|
| B cell activation | GO:0042113 |  | 3.943E-05 | 0.01053966 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 4.111E-05 | 9.892E-03 |
|---|
| thymus development | GO:0048538 |  | 4.561E-05 | 9.975E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 2.097E-08 | 5.124E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.81E-07 | 2.211E-04 |
|---|
| B cell differentiation | GO:0030183 |  | 8.314E-06 | 6.771E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.889E-06 | 5.429E-03 |
|---|
| response to UV | GO:0009411 |  | 1.528E-05 | 7.466E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.054E-05 | 8.365E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.298E-05 | 0.01151093 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.298E-05 | 0.01007206 |
|---|
| B cell activation | GO:0042113 |  | 3.439E-05 | 9.335E-03 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 3.734E-05 | 9.121E-03 |
|---|
| thymus development | GO:0048538 |  | 4.029E-05 | 8.948E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 2.512E-08 | 6.045E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.281E-07 | 2.745E-04 |
|---|
| B cell differentiation | GO:0030183 |  | 9.29E-06 | 7.451E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.064E-05 | 6.397E-03 |
|---|
| response to UV | GO:0009411 |  | 1.634E-05 | 7.863E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.326E-05 | 9.326E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.734E-05 | 0.01283284 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.734E-05 | 0.01122874 |
|---|
| B cell activation | GO:0042113 |  | 3.943E-05 | 0.01053966 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 4.111E-05 | 9.892E-03 |
|---|
| thymus development | GO:0048538 |  | 4.561E-05 | 9.975E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.533E-11 | 1.976E-07 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.73E-08 | 3.089E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.805E-08 | 4.529E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.044E-07 | 9.321E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 2.213E-07 | 1.58E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.618E-06 | 2.748E-03 |
|---|
| bHLH transcription factor binding | GO:0043425 |  | 5.061E-06 | 2.582E-03 |
|---|
| response to UV | GO:0009411 |  | 5.598E-06 | 2.499E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.589E-06 | 3.011E-03 |
|---|
| B cell lineage commitment | GO:0002326 |  | 7.589E-06 | 2.71E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.381E-05 | 4.482E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 9.653E-11 | 3.522E-07 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.125E-08 | 2.052E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.38E-08 | 2.894E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 6.828E-08 | 6.227E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.211E-07 | 8.837E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.448E-06 | 1.488E-03 |
|---|
| response to UV | GO:0009411 |  | 2.813E-06 | 1.466E-03 |
|---|
| bHLH transcription factor binding | GO:0043425 |  | 3.763E-06 | 1.716E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.642E-06 | 2.287E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 8.202E-06 | 2.992E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 1.075E-05 | 3.566E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 2.512E-08 | 6.045E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.281E-07 | 2.745E-04 |
|---|
| B cell differentiation | GO:0030183 |  | 9.29E-06 | 7.451E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.064E-05 | 6.397E-03 |
|---|
| response to UV | GO:0009411 |  | 1.634E-05 | 7.863E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.326E-05 | 9.326E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.734E-05 | 0.01283284 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.734E-05 | 0.01122874 |
|---|
| B cell activation | GO:0042113 |  | 3.943E-05 | 0.01053966 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 4.111E-05 | 9.892E-03 |
|---|
| thymus development | GO:0048538 |  | 4.561E-05 | 9.975E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.018E-07 | 2.342E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.159E-07 | 1.053E-03 |
|---|
| B cell differentiation | GO:0030183 |  | 2.782E-05 | 0.02132696 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.464E-05 | 0.0199204 |
|---|
| response to UV | GO:0009411 |  | 4.249E-05 | 0.01954448 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01704369 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 5.923E-05 | 0.01946068 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 5.923E-05 | 0.01702809 |
|---|
| thymus development | GO:0048538 |  | 7.608E-05 | 0.01944287 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 9.501E-05 | 0.02185322 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 1.146E-04 | 0.02395412 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.018E-07 | 2.342E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.159E-07 | 1.053E-03 |
|---|
| B cell differentiation | GO:0030183 |  | 2.782E-05 | 0.02132696 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.464E-05 | 0.0199204 |
|---|
| response to UV | GO:0009411 |  | 4.249E-05 | 0.01954448 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01704369 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 5.923E-05 | 0.01946068 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 5.923E-05 | 0.01702809 |
|---|
| thymus development | GO:0048538 |  | 7.608E-05 | 0.01944287 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 9.501E-05 | 0.02185322 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 1.146E-04 | 0.02395412 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.018E-07 | 2.342E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.159E-07 | 1.053E-03 |
|---|
| B cell differentiation | GO:0030183 |  | 2.782E-05 | 0.02132696 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.464E-05 | 0.0199204 |
|---|
| response to UV | GO:0009411 |  | 4.249E-05 | 0.01954448 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01704369 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 5.923E-05 | 0.01946068 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 5.923E-05 | 0.01702809 |
|---|
| thymus development | GO:0048538 |  | 7.608E-05 | 0.01944287 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 9.501E-05 | 0.02185322 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 1.146E-04 | 0.02395412 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.018E-07 | 2.342E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.159E-07 | 1.053E-03 |
|---|
| B cell differentiation | GO:0030183 |  | 2.782E-05 | 0.02132696 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.464E-05 | 0.0199204 |
|---|
| response to UV | GO:0009411 |  | 4.249E-05 | 0.01954448 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01704369 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 5.923E-05 | 0.01946068 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 5.923E-05 | 0.01702809 |
|---|
| thymus development | GO:0048538 |  | 7.608E-05 | 0.01944287 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 9.501E-05 | 0.02185322 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 1.146E-04 | 0.02395412 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 6.918E-08 | 1.642E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.433E-07 | 7.632E-04 |
|---|
| B cell differentiation | GO:0030183 |  | 1.898E-05 | 0.01501098 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.535E-05 | 0.01503924 |
|---|
| response to UV | GO:0009411 |  | 3.096E-05 | 0.0146921 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.437E-05 | 0.01359475 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 4.579E-05 | 0.01552436 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 7.348E-05 | 0.02179603 |
|---|
| B cell activation | GO:0042113 |  | 8.594E-05 | 0.02265823 |
|---|
| thymus development | GO:0048538 |  | 8.974E-05 | 0.02129453 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 9.395E-05 | 0.02026811 |
|---|



