Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6628-8-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2291 | 7.709e-03 | 9.197e-03 | 8.141e-02 |
|---|
| IPC-NIBC-129 | 0.2484 | 4.442e-02 | 5.612e-02 | 4.177e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2609 | 7.351e-03 | 1.167e-02 | 2.182e-01 |
|---|
| Loi_GPL570 | 0.2574 | 6.286e-02 | 6.243e-02 | 3.009e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1638 | 2.300e-05 | 1.360e-04 | 2.670e-04 |
|---|
| Parker_GPL1390 | 0.2669 | 6.534e-02 | 6.265e-02 | 4.359e-01 |
|---|
| Parker_GPL887 | 0.0227 | 9.423e-01 | 9.489e-01 | 9.710e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3682 | 9.471e-01 | 9.492e-01 | 1.000e+00 |
|---|
| Schmidt | 0.2272 | 1.286e-02 | 1.817e-02 | 1.584e-01 |
|---|
| Sotiriou | 0.2998 | 4.626e-03 | 1.572e-03 | 9.644e-02 |
|---|
| Wang | 0.1651 | 1.042e-01 | 7.215e-02 | 3.754e-01 |
|---|
| Zhang | 0.0171 | 1.294e-01 | 8.919e-02 | 3.009e-01 |
|---|
| Zhou | 0.3757 | 5.672e-02 | 5.955e-02 | 3.546e-01 |
|---|
Expression data for subnetwork 6628-8-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
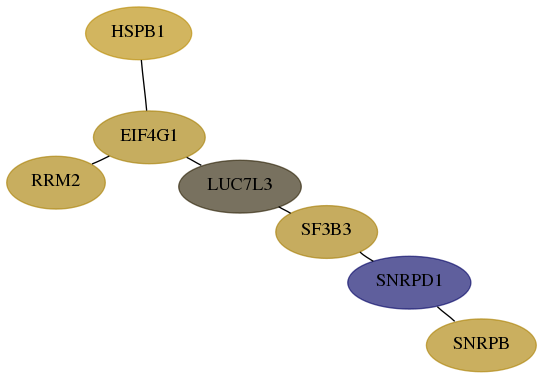
Score for each gene in subnetwork 6628-8-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| snrpb |   | 1 | 164 | 401 | 401 | 0.141 | 0.077 | 0.167 | 0.250 | 0.139 | 0.089 | 0.155 | 0.281 | 0.120 | 0.161 | 0.045 | 0.010 | 0.153 |
|---|
| sf3b3 |   | 7 | 38 | 1 | 5 | 0.119 | 0.241 | 0.143 | -0.019 | 0.082 | 0.222 | 0.007 | 0.271 | 0.168 | 0.060 | 0.017 | -0.077 | 0.372 |
|---|
| rrm2 |   | 21 | 10 | 44 | 26 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | 0.117 | 0.129 | 0.302 |
|---|
| snrpd1 |   | 2 | 104 | 337 | 327 | -0.026 | 0.257 | 0.128 | 0.254 | 0.106 | 0.174 | 0.255 | 0.155 | 0.190 | 0.111 | 0.076 | 0.011 | 0.205 |
|---|
| luc7l3 |   | 7 | 38 | 1 | 5 | 0.006 | 0.111 | -0.046 | -0.016 | 0.121 | 0.071 | -0.318 | -0.084 | 0.159 | 0.098 | 0.155 | 0.067 | 0.065 |
|---|
| eif4g1 |   | 5 | 50 | 153 | 142 | 0.123 | 0.262 | 0.043 | 0.013 | 0.098 | 0.189 | 0.016 | 0.194 | 0.076 | 0.147 | -0.154 | -0.028 | 0.268 |
|---|
GO Enrichment output for subnetwork 6628-8-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.936E-09 | 1.135E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 8.635E-09 | 9.93E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.264E-07 | 2.502E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 3.778E-07 | 2.172E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 3.45E-06 | 1.587E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.054E-05 | 4.04E-03 |
|---|
| regulation of translation | GO:0006417 |  | 3.849E-05 | 0.01264526 |
|---|
| spliceosome assembly | GO:0000245 |  | 1.859E-04 | 0.05344478 |
|---|
| response to heat | GO:0009408 |  | 1.859E-04 | 0.04750647 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 4.021E-04 | 0.09248648 |
|---|
| response to temperature stimulus | GO:0009266 |  | 4.787E-04 | 0.10010159 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 1.245E-07 | 3.043E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 9.378E-07 | 1.145E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.59E-06 | 2.109E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.439E-06 | 2.711E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.111E-05 | 0.01519835 |
|---|
| response to heat | GO:0009408 |  | 5.708E-05 | 0.02323971 |
|---|
| spliceosome assembly | GO:0000245 |  | 6.882E-05 | 0.02401734 |
|---|
| response to temperature stimulus | GO:0009266 |  | 1.559E-04 | 0.04759454 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.621E-04 | 0.04398944 |
|---|
| protein oligomerization | GO:0051259 |  | 3.745E-04 | 0.09147873 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.856E-04 | 0.10785813 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.795E-09 | 6.726E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.705E-09 | 8.067E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.225E-07 | 9.824E-05 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.323E-07 | 1.398E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.848E-06 | 8.891E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 5.959E-06 | 2.39E-03 |
|---|
| response to heat | GO:0009408 |  | 8.664E-05 | 0.0297796 |
|---|
| spliceosome assembly | GO:0000245 |  | 9.874E-05 | 0.02969536 |
|---|
| response to temperature stimulus | GO:0009266 |  | 2.33E-04 | 0.06230183 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 2.33E-04 | 0.05607164 |
|---|
| response to unfolded protein | GO:0006986 |  | 7.007E-04 | 0.15327097 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 1.245E-07 | 3.043E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 9.378E-07 | 1.145E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.59E-06 | 2.109E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.439E-06 | 2.711E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.111E-05 | 0.01519835 |
|---|
| response to heat | GO:0009408 |  | 5.708E-05 | 0.02323971 |
|---|
| spliceosome assembly | GO:0000245 |  | 6.882E-05 | 0.02401734 |
|---|
| response to temperature stimulus | GO:0009266 |  | 1.559E-04 | 0.04759454 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.621E-04 | 0.04398944 |
|---|
| protein oligomerization | GO:0051259 |  | 3.745E-04 | 0.09147873 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.856E-04 | 0.10785813 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.795E-09 | 6.726E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.705E-09 | 8.067E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.225E-07 | 9.824E-05 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.323E-07 | 1.398E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.848E-06 | 8.891E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 5.959E-06 | 2.39E-03 |
|---|
| response to heat | GO:0009408 |  | 8.664E-05 | 0.0297796 |
|---|
| spliceosome assembly | GO:0000245 |  | 9.874E-05 | 0.02969536 |
|---|
| response to temperature stimulus | GO:0009266 |  | 2.33E-04 | 0.06230183 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 2.33E-04 | 0.05607164 |
|---|
| response to unfolded protein | GO:0006986 |  | 7.007E-04 | 0.15327097 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| small nuclear ribonucleoprotein complex | GO:0030532 |  | 2.248E-10 | 8.027E-07 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.387E-07 | 4.262E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 6.946E-07 | 8.268E-04 |
|---|
| oxidoreductase activity. acting on CH or CH2 groups | GO:0016725 |  | 4.058E-06 | 3.623E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.058E-06 | 2.899E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.954E-06 | 4.139E-03 |
|---|
| eukaryotic translation initiation factor 4F complex | GO:0016281 |  | 1.756E-05 | 8.958E-03 |
|---|
| small nucleolar ribonucleoprotein complex | GO:0005732 |  | 2.622E-05 | 0.01170582 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.95E-05 | 0.01170317 |
|---|
| RNA splicing factor activity. transesterification mechanism | GO:0031202 |  | 5.314E-05 | 0.01897465 |
|---|
| response to heat | GO:0009408 |  | 6.254E-05 | 0.02030293 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| small nuclear ribonucleoprotein complex | GO:0030532 |  | 1.449E-10 | 5.286E-07 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 1.491E-07 | 2.72E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 4.291E-07 | 5.218E-04 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.395E-06 | 2.184E-03 |
|---|
| oxidoreductase activity. acting on CH or CH2 groups | GO:0016725 |  | 3.352E-06 | 2.446E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.469E-06 | 2.717E-03 |
|---|
| eukaryotic translation initiation factor 4F complex | GO:0016281 |  | 4.469E-06 | 2.329E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.723E-05 | 0.01241602 |
|---|
| RNA splicing factor activity. transesterification mechanism | GO:0031202 |  | 4.39E-05 | 0.01779421 |
|---|
| response to heat | GO:0009408 |  | 6.007E-05 | 0.02191455 |
|---|
| spliceosome assembly | GO:0000245 |  | 6.91E-05 | 0.02291667 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.795E-09 | 6.726E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.705E-09 | 8.067E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.225E-07 | 9.824E-05 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.323E-07 | 1.398E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.848E-06 | 8.891E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 5.959E-06 | 2.39E-03 |
|---|
| response to heat | GO:0009408 |  | 8.664E-05 | 0.0297796 |
|---|
| spliceosome assembly | GO:0000245 |  | 9.874E-05 | 0.02969536 |
|---|
| response to temperature stimulus | GO:0009266 |  | 2.33E-04 | 0.06230183 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 2.33E-04 | 0.05607164 |
|---|
| response to unfolded protein | GO:0006986 |  | 7.007E-04 | 0.15327097 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.936E-09 | 1.135E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 8.635E-09 | 9.93E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.264E-07 | 2.502E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 3.778E-07 | 2.172E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 3.45E-06 | 1.587E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.054E-05 | 4.04E-03 |
|---|
| regulation of translation | GO:0006417 |  | 3.849E-05 | 0.01264526 |
|---|
| spliceosome assembly | GO:0000245 |  | 1.859E-04 | 0.05344478 |
|---|
| response to heat | GO:0009408 |  | 1.859E-04 | 0.04750647 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 4.021E-04 | 0.09248648 |
|---|
| response to temperature stimulus | GO:0009266 |  | 4.787E-04 | 0.10010159 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.936E-09 | 1.135E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 8.635E-09 | 9.93E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.264E-07 | 2.502E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 3.778E-07 | 2.172E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 3.45E-06 | 1.587E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.054E-05 | 4.04E-03 |
|---|
| regulation of translation | GO:0006417 |  | 3.849E-05 | 0.01264526 |
|---|
| spliceosome assembly | GO:0000245 |  | 1.859E-04 | 0.05344478 |
|---|
| response to heat | GO:0009408 |  | 1.859E-04 | 0.04750647 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 4.021E-04 | 0.09248648 |
|---|
| response to temperature stimulus | GO:0009266 |  | 4.787E-04 | 0.10010159 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.936E-09 | 1.135E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 8.635E-09 | 9.93E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.264E-07 | 2.502E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 3.778E-07 | 2.172E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 3.45E-06 | 1.587E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.054E-05 | 4.04E-03 |
|---|
| regulation of translation | GO:0006417 |  | 3.849E-05 | 0.01264526 |
|---|
| spliceosome assembly | GO:0000245 |  | 1.859E-04 | 0.05344478 |
|---|
| response to heat | GO:0009408 |  | 1.859E-04 | 0.04750647 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 4.021E-04 | 0.09248648 |
|---|
| response to temperature stimulus | GO:0009266 |  | 4.787E-04 | 0.10010159 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.936E-09 | 1.135E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 8.635E-09 | 9.93E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.264E-07 | 2.502E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 3.778E-07 | 2.172E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 3.45E-06 | 1.587E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 1.054E-05 | 4.04E-03 |
|---|
| regulation of translation | GO:0006417 |  | 3.849E-05 | 0.01264526 |
|---|
| spliceosome assembly | GO:0000245 |  | 1.859E-04 | 0.05344478 |
|---|
| response to heat | GO:0009408 |  | 1.859E-04 | 0.04750647 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 4.021E-04 | 0.09248648 |
|---|
| response to temperature stimulus | GO:0009266 |  | 4.787E-04 | 0.10010159 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 1.91E-09 | 4.531E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 7.625E-09 | 9.047E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.219E-08 | 9.646E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.336E-07 | 1.979E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 3.259E-06 | 1.547E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 9.749E-06 | 3.856E-03 |
|---|
| response to heat | GO:0009408 |  | 1.698E-04 | 0.05756633 |
|---|
| spliceosome assembly | GO:0000245 |  | 2.454E-04 | 0.07278921 |
|---|
| response to temperature stimulus | GO:0009266 |  | 4.559E-04 | 0.12019865 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 5.537E-04 | 0.13139349 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.211E-03 | 0.26125594 |
|---|



