Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6119-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2116 | 5.971e-03 | 7.109e-03 | 6.693e-02 |
|---|
| IPC-NIBC-129 | 0.2068 | 4.070e-02 | 5.112e-02 | 3.929e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2732 | 1.326e-02 | 1.985e-02 | 3.106e-01 |
|---|
| Loi_GPL570 | 0.2297 | 1.100e-01 | 1.094e-01 | 4.399e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1736 | 5.000e-06 | 3.300e-05 | 2.300e-05 |
|---|
| Parker_GPL1390 | 0.2732 | 8.357e-02 | 8.199e-02 | 4.951e-01 |
|---|
| Parker_GPL887 | 0.1501 | 4.851e-01 | 5.190e-01 | 5.183e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3488 | 2.749e-01 | 2.356e-01 | 8.414e-01 |
|---|
| Schmidt | 0.2500 | 8.353e-03 | 1.203e-02 | 1.136e-01 |
|---|
| Sotiriou | 0.3261 | 3.294e-03 | 1.025e-03 | 7.596e-02 |
|---|
| Wang | 0.2181 | 8.898e-02 | 6.000e-02 | 3.385e-01 |
|---|
| Zhang | 0.1698 | 6.040e-02 | 3.581e-02 | 1.519e-01 |
|---|
| Zhou | 0.3962 | 6.824e-02 | 7.290e-02 | 4.142e-01 |
|---|
Expression data for subnetwork 6119-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
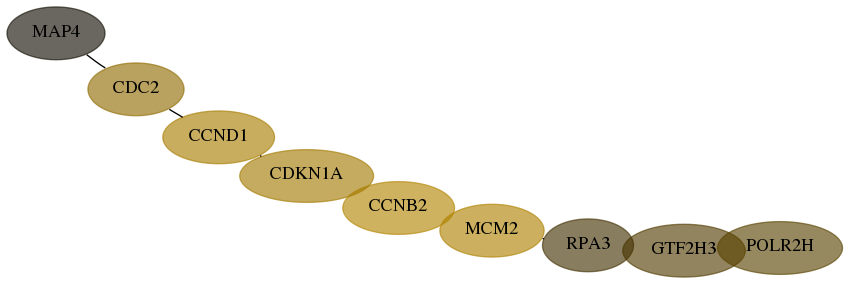
Score for each gene in subnetwork 6119-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| gtf2h3 |   | 1 | 164 | 216 | 229 | 0.016 | 0.083 | -0.028 | 0.080 | 0.062 | 0.157 | -0.175 | -0.119 | 0.106 | 0.100 | 0.107 | 0.002 | -0.162 |
|---|
| ccnb2 |   | 16 | 13 | 153 | 125 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.178 | 0.090 | 0.366 | 0.240 | 0.169 | 0.144 | 0.211 | 0.463 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| mcm2 |   | 4 | 60 | 216 | 192 | 0.151 | 0.254 | 0.188 | 0.128 | 0.154 | 0.220 | -0.034 | 0.267 | 0.224 | 0.200 | 0.101 | 0.012 | 0.255 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| polr2h |   | 3 | 79 | 175 | 162 | 0.021 | 0.282 | 0.144 | 0.302 | 0.127 | 0.204 | 0.069 | 0.129 | 0.257 | 0.153 | 0.128 | 0.165 | 0.236 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| map4 |   | 16 | 13 | 73 | 55 | 0.002 | 0.046 | 0.067 | -0.043 | 0.034 | 0.119 | 0.159 | -0.058 | -0.020 | 0.043 | 0.108 | -0.074 | 0.302 |
|---|
| rpa3 |   | 2 | 104 | 216 | 208 | 0.011 | 0.107 | 0.109 | 0.001 | 0.162 | 0.044 | -0.259 | 0.191 | 0.126 | 0.132 | 0.149 | 0.043 | 0.108 |
|---|
GO Enrichment output for subnetwork 6119-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.005E-06 | 2.311E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 0.01058205 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 8.662E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.206E-05 | 6.935E-03 |
|---|
| RNA elongation | GO:0006354 |  | 1.369E-05 | 6.296E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.545E-05 | 5.924E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.639E-05 | 5.385E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.837E-05 | 5.282E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.763E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.484E-05 | 5.712E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.484E-05 | 5.193E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 3.615E-07 | 8.832E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.07E-06 | 3.75E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 4.459E-06 | 3.631E-03 |
|---|
| response to UV | GO:0009411 |  | 5.287E-06 | 3.229E-03 |
|---|
| RNA elongation | GO:0006354 |  | 5.584E-06 | 2.729E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.892E-06 | 2.399E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 5.892E-06 | 2.056E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 6.883E-06 | 2.102E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.022E-05 | 2.773E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.641E-05 | 4.008E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 1.641E-05 | 3.644E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 5.128E-07 | 1.234E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-06 | 5.233E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 5.955E-06 | 4.776E-03 |
|---|
| response to UV | GO:0009411 |  | 6.692E-06 | 4.025E-03 |
|---|
| RNA elongation | GO:0006354 |  | 7.488E-06 | 3.603E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.908E-06 | 3.171E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.344E-06 | 2.868E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 9.262E-06 | 2.786E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.293E-05 | 3.457E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.077E-05 | 4.997E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.077E-05 | 4.542E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 3.615E-07 | 8.832E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.07E-06 | 3.75E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 4.459E-06 | 3.631E-03 |
|---|
| response to UV | GO:0009411 |  | 5.287E-06 | 3.229E-03 |
|---|
| RNA elongation | GO:0006354 |  | 5.584E-06 | 2.729E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.892E-06 | 2.399E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 5.892E-06 | 2.056E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 6.883E-06 | 2.102E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.022E-05 | 2.773E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.641E-05 | 4.008E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 1.641E-05 | 3.644E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 5.128E-07 | 1.234E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-06 | 5.233E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 5.955E-06 | 4.776E-03 |
|---|
| response to UV | GO:0009411 |  | 6.692E-06 | 4.025E-03 |
|---|
| RNA elongation | GO:0006354 |  | 7.488E-06 | 3.603E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.908E-06 | 3.171E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.344E-06 | 2.868E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 9.262E-06 | 2.786E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.293E-05 | 3.457E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.077E-05 | 4.997E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.077E-05 | 4.542E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 5.876E-09 | 2.098E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.73E-08 | 3.089E-05 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.851E-07 | 2.204E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.578E-06 | 1.408E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.578E-06 | 1.127E-03 |
|---|
| RNA elongation | GO:0006354 |  | 1.913E-06 | 1.139E-03 |
|---|
| response to UV | GO:0009411 |  | 1.913E-06 | 9.76E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.035E-06 | 9.083E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 2.572E-06 | 1.021E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.72E-06 | 9.714E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.865E-06 | 1.255E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 4.298E-09 | 1.568E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 2.79E-08 | 5.088E-05 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.018E-07 | 1.237E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.392E-07 | 8.565E-04 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.008E-06 | 7.352E-04 |
|---|
| response to UV | GO:0009411 |  | 1.079E-06 | 6.563E-04 |
|---|
| RNA elongation | GO:0006354 |  | 1.233E-06 | 6.425E-04 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.4E-06 | 6.384E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.582E-06 | 6.412E-04 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.779E-06 | 6.488E-04 |
|---|
| origin recognition complex | GO:0000808 |  | 2.053E-06 | 6.809E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 5.128E-07 | 1.234E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-06 | 5.233E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 5.955E-06 | 4.776E-03 |
|---|
| response to UV | GO:0009411 |  | 6.692E-06 | 4.025E-03 |
|---|
| RNA elongation | GO:0006354 |  | 7.488E-06 | 3.603E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.908E-06 | 3.171E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.344E-06 | 2.868E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 9.262E-06 | 2.786E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.293E-05 | 3.457E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.077E-05 | 4.997E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.077E-05 | 4.542E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.005E-06 | 2.311E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 0.01058205 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 8.662E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.206E-05 | 6.935E-03 |
|---|
| RNA elongation | GO:0006354 |  | 1.369E-05 | 6.296E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.545E-05 | 5.924E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.639E-05 | 5.385E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.837E-05 | 5.282E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.763E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.484E-05 | 5.712E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.484E-05 | 5.193E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.005E-06 | 2.311E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 0.01058205 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 8.662E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.206E-05 | 6.935E-03 |
|---|
| RNA elongation | GO:0006354 |  | 1.369E-05 | 6.296E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.545E-05 | 5.924E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.639E-05 | 5.385E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.837E-05 | 5.282E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.763E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.484E-05 | 5.712E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.484E-05 | 5.193E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.005E-06 | 2.311E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 0.01058205 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 8.662E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.206E-05 | 6.935E-03 |
|---|
| RNA elongation | GO:0006354 |  | 1.369E-05 | 6.296E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.545E-05 | 5.924E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.639E-05 | 5.385E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.837E-05 | 5.282E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.763E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.484E-05 | 5.712E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.484E-05 | 5.193E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.005E-06 | 2.311E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 0.01058205 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 8.662E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.206E-05 | 6.935E-03 |
|---|
| RNA elongation | GO:0006354 |  | 1.369E-05 | 6.296E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.545E-05 | 5.924E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.639E-05 | 5.385E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.837E-05 | 5.282E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.763E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.484E-05 | 5.712E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.484E-05 | 5.193E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 7.909E-07 | 1.877E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.723E-06 | 7.977E-03 |
|---|
| response to UV | GO:0009411 |  | 8.218E-06 | 6.501E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 9.328E-06 | 5.534E-03 |
|---|
| RNA elongation | GO:0006354 |  | 1.053E-05 | 4.999E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.184E-05 | 4.681E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.252E-05 | 4.246E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.398E-05 | 4.148E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.441E-05 | 3.798E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.92E-05 | 4.556E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.082E-05 | 6.649E-03 |
|---|



