Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 51022-5-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2208 | 4.322e-03 | 5.129e-03 | 5.204e-02 |
|---|
| IPC-NIBC-129 | 0.2780 | 3.528e-02 | 4.387e-02 | 3.544e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2888 | 1.672e-02 | 2.445e-02 | 3.535e-01 |
|---|
| Loi_GPL570 | 0.2188 | 4.187e-02 | 4.154e-02 | 2.232e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1650 | 9.000e-06 | 6.000e-05 | 6.600e-05 |
|---|
| Parker_GPL1390 | 0.2835 | 7.343e-02 | 7.119e-02 | 4.634e-01 |
|---|
| Parker_GPL887 | 0.1630 | 4.485e-01 | 4.831e-01 | 4.762e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3435 | 6.129e-01 | 5.889e-01 | 9.884e-01 |
|---|
| Schmidt | 0.2405 | 7.262e-03 | 1.052e-02 | 1.017e-01 |
|---|
| Sotiriou | 0.3346 | 3.943e-03 | 1.286e-03 | 8.626e-02 |
|---|
| Wang | 0.2061 | 1.021e-01 | 7.049e-02 | 3.706e-01 |
|---|
| Zhang | 0.0806 | 7.192e-02 | 4.414e-02 | 1.785e-01 |
|---|
| Zhou | 0.3854 | 7.175e-02 | 7.700e-02 | 4.312e-01 |
|---|
Expression data for subnetwork 51022-5-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
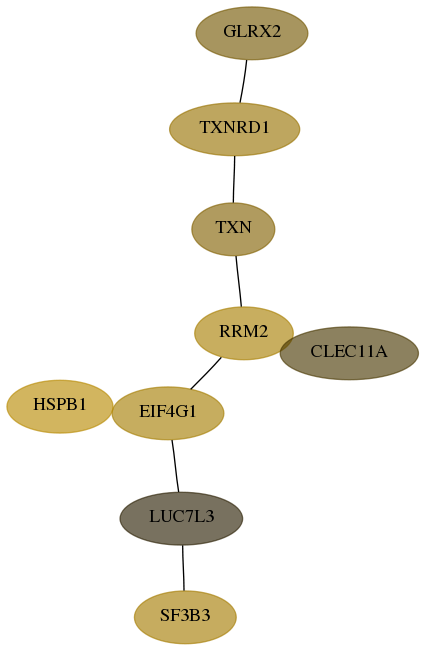
Score for each gene in subnetwork 51022-5-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| rrm2 |   | 21 | 10 | 44 | 26 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | 0.117 | 0.129 | 0.302 |
|---|
| luc7l3 |   | 7 | 38 | 1 | 5 | 0.006 | 0.111 | -0.046 | -0.016 | 0.121 | 0.071 | -0.318 | -0.084 | 0.159 | 0.098 | 0.155 | 0.067 | 0.065 |
|---|
| eif4g1 |   | 5 | 50 | 153 | 142 | 0.123 | 0.262 | 0.043 | 0.013 | 0.098 | 0.189 | 0.016 | 0.194 | 0.076 | 0.147 | -0.154 | -0.028 | 0.268 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| txn |   | 1 | 164 | 244 | 247 | 0.052 | 0.168 | 0.117 | 0.209 | 0.171 | 0.155 | 0.166 | 0.246 | 0.161 | 0.168 | 0.115 | 0.132 | 0.324 |
|---|
| sf3b3 |   | 7 | 38 | 1 | 5 | 0.119 | 0.241 | 0.143 | -0.019 | 0.082 | 0.222 | 0.007 | 0.271 | 0.168 | 0.060 | 0.017 | -0.077 | 0.372 |
|---|
| txnrd1 |   | 1 | 164 | 244 | 247 | 0.090 | 0.133 | 0.124 | 0.256 | 0.156 | 0.162 | 0.204 | 0.101 | 0.169 | 0.142 | 0.155 | 0.118 | 0.215 |
|---|
| clec11a |   | 17 | 12 | 44 | 27 | 0.013 | 0.101 | 0.071 | -0.085 | 0.066 | 0.082 | 0.209 | -0.126 | -0.034 | -0.001 | 0.191 | 0.059 | -0.075 |
|---|
| glrx2 |   | 1 | 164 | 244 | 247 | 0.038 | 0.231 | 0.122 | 0.021 | 0.152 | -0.160 | 0.135 | 0.284 | 0.131 | 0.171 | 0.127 | 0.070 | 0.130 |
|---|
GO Enrichment output for subnetwork 51022-5-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.256E-08 | 2.888E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 2.197E-08 | 2.526E-05 |
|---|
| cell redox homeostasis | GO:0045454 |  | 7.46E-08 | 5.719E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 3.063E-07 | 1.761E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.282E-07 | 3.81E-04 |
|---|
| electron transport chain | GO:0022900 |  | 1.358E-06 | 5.204E-04 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 2.265E-06 | 7.443E-04 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 4.409E-06 | 1.268E-03 |
|---|
| protein thiol-disulfide exchange | GO:0006467 |  | 7.841E-06 | 2.004E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 8.719E-06 | 2.005E-03 |
|---|
| response to temperature stimulus | GO:0009266 |  | 1.061E-05 | 2.218E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell redox homeostasis | GO:0045454 |  | 1.245E-12 | 3.043E-09 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 6.781E-12 | 8.283E-09 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 2.638E-11 | 2.148E-08 |
|---|
| cellular response to hydrogen peroxide | GO:0070301 |  | 2.248E-10 | 1.373E-07 |
|---|
| cellular response to reactive oxygen species | GO:0034614 |  | 5.129E-10 | 2.506E-07 |
|---|
| hydrogen peroxide metabolic process | GO:0042743 |  | 8.65E-10 | 3.522E-07 |
|---|
| cellular response to oxidative stress | GO:0034599 |  | 1.015E-09 | 3.543E-07 |
|---|
| mesoderm formation | GO:0001707 |  | 2.076E-09 | 6.341E-07 |
|---|
| formation of primary germ layer | GO:0001704 |  | 2.676E-09 | 7.264E-07 |
|---|
| mesoderm morphogenesis | GO:0048332 |  | 3.02E-09 | 7.379E-07 |
|---|
| gastrulation | GO:0007369 |  | 2.428E-08 | 5.393E-06 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell redox homeostasis | GO:0045454 |  | 4.198E-10 | 1.01E-06 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 4.022E-09 | 4.838E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 7.438E-09 | 5.965E-06 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 1.247E-08 | 7.5E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.784E-08 | 8.583E-06 |
|---|
| protein oligomerization | GO:0051259 |  | 1.566E-07 | 6.281E-05 |
|---|
| cellular response to hydrogen peroxide | GO:0070301 |  | 1.727E-07 | 5.934E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.253E-07 | 9.784E-05 |
|---|
| cellular response to reactive oxygen species | GO:0034614 |  | 3.253E-07 | 8.697E-05 |
|---|
| hydrogen peroxide metabolic process | GO:0042743 |  | 4.853E-07 | 1.168E-04 |
|---|
| cellular response to oxidative stress | GO:0034599 |  | 5.484E-07 | 1.199E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell redox homeostasis | GO:0045454 |  | 1.245E-12 | 3.043E-09 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 6.781E-12 | 8.283E-09 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 2.638E-11 | 2.148E-08 |
|---|
| cellular response to hydrogen peroxide | GO:0070301 |  | 2.248E-10 | 1.373E-07 |
|---|
| cellular response to reactive oxygen species | GO:0034614 |  | 5.129E-10 | 2.506E-07 |
|---|
| hydrogen peroxide metabolic process | GO:0042743 |  | 8.65E-10 | 3.522E-07 |
|---|
| cellular response to oxidative stress | GO:0034599 |  | 1.015E-09 | 3.543E-07 |
|---|
| mesoderm formation | GO:0001707 |  | 2.076E-09 | 6.341E-07 |
|---|
| formation of primary germ layer | GO:0001704 |  | 2.676E-09 | 7.264E-07 |
|---|
| mesoderm morphogenesis | GO:0048332 |  | 3.02E-09 | 7.379E-07 |
|---|
| gastrulation | GO:0007369 |  | 2.428E-08 | 5.393E-06 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell redox homeostasis | GO:0045454 |  | 4.198E-10 | 1.01E-06 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 4.022E-09 | 4.838E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 7.438E-09 | 5.965E-06 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 1.247E-08 | 7.5E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.784E-08 | 8.583E-06 |
|---|
| protein oligomerization | GO:0051259 |  | 1.566E-07 | 6.281E-05 |
|---|
| cellular response to hydrogen peroxide | GO:0070301 |  | 1.727E-07 | 5.934E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.253E-07 | 9.784E-05 |
|---|
| cellular response to reactive oxygen species | GO:0034614 |  | 3.253E-07 | 8.697E-05 |
|---|
| hydrogen peroxide metabolic process | GO:0042743 |  | 4.853E-07 | 1.168E-04 |
|---|
| cellular response to oxidative stress | GO:0034599 |  | 5.484E-07 | 1.199E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell redox homeostasis | GO:0045454 |  | 1.46E-08 | 5.213E-05 |
|---|
| protein disulfide oxidoreductase activity | GO:0015035 |  | 3.458E-08 | 6.174E-05 |
|---|
| disulfide oxidoreductase activity | GO:0015036 |  | 7.145E-08 | 8.504E-05 |
|---|
| electron transport chain | GO:0022900 |  | 2.452E-07 | 2.189E-04 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 7.748E-07 | 5.534E-04 |
|---|
| oxidoreductase activity. acting on sulfur group of donors | GO:0016667 |  | 9.342E-07 | 5.56E-04 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 1.661E-06 | 8.472E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.661E-06 | 7.413E-04 |
|---|
| response to temperature stimulus | GO:0009266 |  | 2.208E-06 | 8.76E-04 |
|---|
| protein thiol-disulfide exchange | GO:0006467 |  | 4.969E-06 | 1.774E-03 |
|---|
| thioredoxin-disulfide reductase activity | GO:0004791 |  | 4.969E-06 | 1.613E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell redox homeostasis | GO:0045454 |  | 1.098E-08 | 4.005E-05 |
|---|
| protein disulfide oxidoreductase activity | GO:0015035 |  | 1.807E-08 | 3.296E-05 |
|---|
| disulfide oxidoreductase activity | GO:0015036 |  | 4.978E-08 | 6.053E-05 |
|---|
| electron transport chain | GO:0022900 |  | 1.389E-07 | 1.267E-04 |
|---|
| oxidoreductase activity. acting on sulfur group of donors | GO:0016667 |  | 5.939E-07 | 4.333E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 9.17E-07 | 5.575E-04 |
|---|
| response to temperature stimulus | GO:0009266 |  | 1.996E-06 | 1.04E-03 |
|---|
| protein thiol-disulfide exchange | GO:0006467 |  | 3.848E-06 | 1.755E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.848E-06 | 1.56E-03 |
|---|
| oxidoreductase activity. acting on CH or CH2 groups | GO:0016725 |  | 5.386E-06 | 1.965E-03 |
|---|
| response to redox state | GO:0051775 |  | 5.386E-06 | 1.786E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell redox homeostasis | GO:0045454 |  | 4.198E-10 | 1.01E-06 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 4.022E-09 | 4.838E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 7.438E-09 | 5.965E-06 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 1.247E-08 | 7.5E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.784E-08 | 8.583E-06 |
|---|
| protein oligomerization | GO:0051259 |  | 1.566E-07 | 6.281E-05 |
|---|
| cellular response to hydrogen peroxide | GO:0070301 |  | 1.727E-07 | 5.934E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.253E-07 | 9.784E-05 |
|---|
| cellular response to reactive oxygen species | GO:0034614 |  | 3.253E-07 | 8.697E-05 |
|---|
| hydrogen peroxide metabolic process | GO:0042743 |  | 4.853E-07 | 1.168E-04 |
|---|
| cellular response to oxidative stress | GO:0034599 |  | 5.484E-07 | 1.199E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.256E-08 | 2.888E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 2.197E-08 | 2.526E-05 |
|---|
| cell redox homeostasis | GO:0045454 |  | 7.46E-08 | 5.719E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 3.063E-07 | 1.761E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.282E-07 | 3.81E-04 |
|---|
| electron transport chain | GO:0022900 |  | 1.358E-06 | 5.204E-04 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 2.265E-06 | 7.443E-04 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 4.409E-06 | 1.268E-03 |
|---|
| protein thiol-disulfide exchange | GO:0006467 |  | 7.841E-06 | 2.004E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 8.719E-06 | 2.005E-03 |
|---|
| response to temperature stimulus | GO:0009266 |  | 1.061E-05 | 2.218E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.256E-08 | 2.888E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 2.197E-08 | 2.526E-05 |
|---|
| cell redox homeostasis | GO:0045454 |  | 7.46E-08 | 5.719E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 3.063E-07 | 1.761E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.282E-07 | 3.81E-04 |
|---|
| electron transport chain | GO:0022900 |  | 1.358E-06 | 5.204E-04 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 2.265E-06 | 7.443E-04 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 4.409E-06 | 1.268E-03 |
|---|
| protein thiol-disulfide exchange | GO:0006467 |  | 7.841E-06 | 2.004E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 8.719E-06 | 2.005E-03 |
|---|
| response to temperature stimulus | GO:0009266 |  | 1.061E-05 | 2.218E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.256E-08 | 2.888E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 2.197E-08 | 2.526E-05 |
|---|
| cell redox homeostasis | GO:0045454 |  | 7.46E-08 | 5.719E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 3.063E-07 | 1.761E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.282E-07 | 3.81E-04 |
|---|
| electron transport chain | GO:0022900 |  | 1.358E-06 | 5.204E-04 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 2.265E-06 | 7.443E-04 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 4.409E-06 | 1.268E-03 |
|---|
| protein thiol-disulfide exchange | GO:0006467 |  | 7.841E-06 | 2.004E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 8.719E-06 | 2.005E-03 |
|---|
| response to temperature stimulus | GO:0009266 |  | 1.061E-05 | 2.218E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.256E-08 | 2.888E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 2.197E-08 | 2.526E-05 |
|---|
| cell redox homeostasis | GO:0045454 |  | 7.46E-08 | 5.719E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 3.063E-07 | 1.761E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.282E-07 | 3.81E-04 |
|---|
| electron transport chain | GO:0022900 |  | 1.358E-06 | 5.204E-04 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 2.265E-06 | 7.443E-04 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 4.409E-06 | 1.268E-03 |
|---|
| protein thiol-disulfide exchange | GO:0006467 |  | 7.841E-06 | 2.004E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 8.719E-06 | 2.005E-03 |
|---|
| response to temperature stimulus | GO:0009266 |  | 1.061E-05 | 2.218E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.492E-08 | 3.541E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 2.386E-08 | 2.832E-05 |
|---|
| cell redox homeostasis | GO:0045454 |  | 6.291E-08 | 4.976E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 1.953E-07 | 1.159E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 6.518E-07 | 3.093E-04 |
|---|
| electron transport chain | GO:0022900 |  | 1.004E-06 | 3.971E-04 |
|---|
| response to hydrogen peroxide | GO:0042542 |  | 1.894E-06 | 6.421E-04 |
|---|
| response to reactive oxygen species | GO:0000302 |  | 3.836E-06 | 1.138E-03 |
|---|
| protein thiol-disulfide exchange | GO:0006467 |  | 6.06E-06 | 1.598E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 6.35E-06 | 1.507E-03 |
|---|
| response to temperature stimulus | GO:0009266 |  | 7.695E-06 | 1.66E-03 |
|---|



