Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 369-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2595 | 1.325e-02 | 1.586e-02 | 1.221e-01 |
|---|
| IPC-NIBC-129 | 0.2515 | 1.172e-02 | 1.326e-02 | 1.403e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2346 | 2.832e-03 | 4.950e-03 | 1.160e-01 |
|---|
| Loi_GPL570 | 0.3017 | 6.026e-02 | 5.984e-02 | 2.920e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1355 | 1.000e-06 | 1.100e-05 | 3.000e-06 |
|---|
| Parker_GPL1390 | 0.3620 | 4.241e-02 | 3.896e-02 | 3.434e-01 |
|---|
| Parker_GPL887 | 0.0893 | 6.885e-01 | 7.147e-01 | 7.432e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3669 | 1.549e-01 | 1.214e-01 | 6.608e-01 |
|---|
| Schmidt | 0.2673 | 7.425e-03 | 1.074e-02 | 1.035e-01 |
|---|
| Sotiriou | 0.3333 | 8.033e-03 | 3.150e-03 | 1.405e-01 |
|---|
| Wang | 0.1686 | 1.635e-01 | 1.221e-01 | 4.971e-01 |
|---|
| Zhang | 0.2281 | 1.231e-01 | 8.402e-02 | 2.882e-01 |
|---|
| Zhou | 0.3422 | 5.741e-02 | 6.034e-02 | 3.583e-01 |
|---|
Expression data for subnetwork 369-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
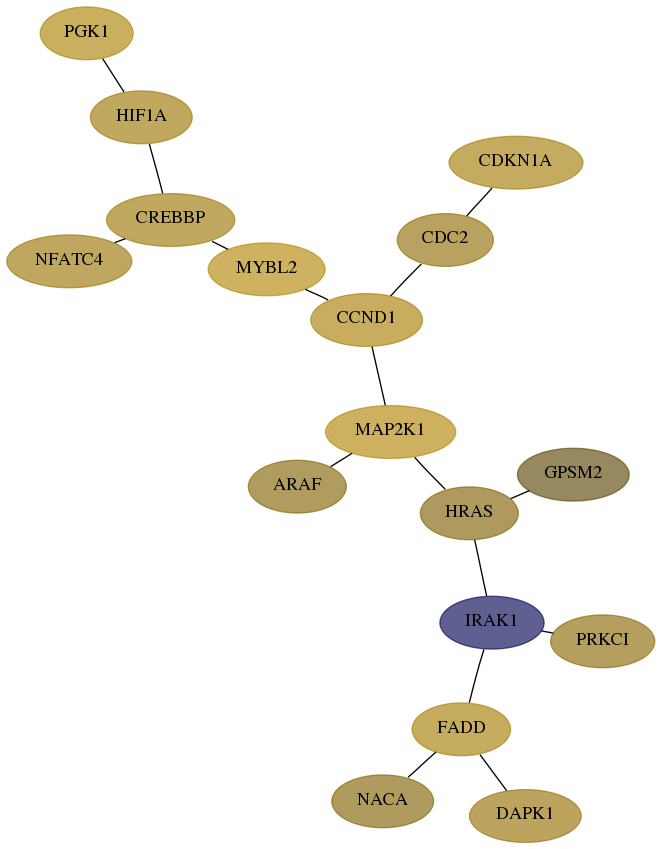
Score for each gene in subnetwork 369-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| irak1 |   | 3 | 79 | 120 | 112 | -0.016 | 0.242 | 0.007 | 0.133 | 0.108 | 0.303 | 0.257 | 0.133 | 0.177 | 0.084 | -0.046 | -0.026 | 0.081 |
|---|
| crebbp |   | 2 | 104 | 120 | 118 | 0.097 | 0.021 | -0.123 | -0.192 | 0.015 | 0.065 | 0.145 | -0.245 | -0.055 | -0.054 | 0.153 | 0.086 | 0.300 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| prkci |   | 12 | 20 | 31 | 19 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | -0.054 | 0.124 | 0.149 |
|---|
| hif1a |   | 2 | 104 | 120 | 118 | 0.097 | 0.246 | 0.089 | -0.009 | 0.101 | 0.088 | -0.152 | 0.011 | -0.043 | 0.119 | 0.172 | 0.129 | -0.135 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| hras |   | 7 | 38 | 88 | 75 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.195 | 0.022 | 0.245 | 0.086 | 0.208 | 0.001 | 0.003 | 0.197 |
|---|
| map2k1 |   | 2 | 104 | 120 | 118 | 0.154 | 0.096 | 0.093 | 0.013 | 0.095 | 0.045 | -0.223 | 0.207 | 0.082 | 0.096 | -0.043 | undef | 0.230 |
|---|
| mybl2 |   | 3 | 79 | 44 | 44 | 0.162 | 0.237 | 0.166 | 0.079 | 0.124 | 0.184 | 0.333 | 0.275 | 0.202 | 0.182 | 0.120 | 0.074 | 0.245 |
|---|
| araf |   | 1 | 164 | 120 | 143 | 0.055 | -0.065 | 0.131 | 0.017 | 0.080 | 0.133 | 0.377 | -0.081 | 0.030 | -0.050 | -0.129 | -0.124 | 0.167 |
|---|
| nfatc4 |   | 8 | 33 | 31 | 25 | 0.094 | -0.056 | -0.021 | 0.204 | 0.079 | -0.008 | 0.243 | 0.060 | 0.077 | -0.012 | 0.080 | 0.131 | 0.154 |
|---|
| pgk1 |   | 5 | 50 | 53 | 47 | 0.141 | 0.293 | 0.135 | 0.197 | 0.170 | 0.170 | 0.120 | 0.310 | 0.201 | 0.127 | 0.110 | 0.054 | 0.141 |
|---|
| fadd |   | 5 | 50 | 64 | 58 | 0.121 | -0.009 | 0.155 | 0.307 | 0.166 | 0.119 | 0.084 | 0.300 | 0.070 | 0.116 | 0.100 | 0.032 | 0.243 |
|---|
| gpsm2 |   | 4 | 60 | 120 | 108 | 0.021 | 0.286 | 0.155 | 0.144 | 0.108 | 0.242 | 0.307 | 0.134 | 0.234 | 0.077 | 0.176 | 0.098 | 0.207 |
|---|
| dapk1 |   | 3 | 79 | 120 | 112 | 0.082 | 0.188 | -0.078 | -0.063 | 0.067 | 0.083 | -0.195 | 0.267 | 0.117 | 0.059 | -0.061 | 0.142 | 0.230 |
|---|
| naca |   | 4 | 60 | 120 | 108 | 0.052 | 0.141 | 0.119 | 0.132 | 0.102 | -0.017 | -0.128 | 0.081 | 0.076 | 0.100 | 0.129 | 0.147 | 0.138 |
|---|
GO Enrichment output for subnetwork 369-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 4.095E-08 | 9.418E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.424E-07 | 3.938E-04 |
|---|
| glycolysis | GO:0006096 |  | 7.916E-07 | 6.069E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.823E-06 | 1.048E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.621E-06 | 1.666E-03 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.494E-06 | 2.106E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 8.851E-06 | 2.908E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.61E-05 | 4.628E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.825E-05 | 4.665E-03 |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 2.649E-05 | 6.092E-03 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 2.649E-05 | 5.538E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.037E-08 | 2.533E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.571E-07 | 1.919E-04 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.703E-07 | 1.387E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 3.51E-07 | 2.144E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 7.713E-07 | 3.769E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.273E-06 | 5.184E-04 |
|---|
| alcohol catabolic process | GO:0046164 |  | 2.079E-06 | 7.255E-04 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 1.052E-05 | 3.213E-03 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.052E-05 | 2.856E-03 |
|---|
| mRNA transcription | GO:0009299 |  | 1.052E-05 | 2.571E-03 |
|---|
| positive regulation of chemokine production | GO:0032722 |  | 1.052E-05 | 2.337E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.566E-08 | 3.767E-05 |
|---|
| glycolysis | GO:0006096 |  | 2.473E-07 | 2.975E-04 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.571E-07 | 2.062E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 5.613E-07 | 3.376E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.249E-06 | 6.008E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.074E-06 | 8.317E-04 |
|---|
| alcohol catabolic process | GO:0046164 |  | 3.404E-06 | 1.17E-03 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 1.386E-05 | 4.169E-03 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.386E-05 | 3.705E-03 |
|---|
| mRNA transcription | GO:0009299 |  | 1.386E-05 | 3.335E-03 |
|---|
| positive regulation of chemokine production | GO:0032722 |  | 1.386E-05 | 3.032E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.037E-08 | 2.533E-05 |
|---|
| glycolysis | GO:0006096 |  | 1.571E-07 | 1.919E-04 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.703E-07 | 1.387E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 3.51E-07 | 2.144E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 7.713E-07 | 3.769E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.273E-06 | 5.184E-04 |
|---|
| alcohol catabolic process | GO:0046164 |  | 2.079E-06 | 7.255E-04 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 1.052E-05 | 3.213E-03 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.052E-05 | 2.856E-03 |
|---|
| mRNA transcription | GO:0009299 |  | 1.052E-05 | 2.571E-03 |
|---|
| positive regulation of chemokine production | GO:0032722 |  | 1.052E-05 | 2.337E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.566E-08 | 3.767E-05 |
|---|
| glycolysis | GO:0006096 |  | 2.473E-07 | 2.975E-04 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.571E-07 | 2.062E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 5.613E-07 | 3.376E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.249E-06 | 6.008E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.074E-06 | 8.317E-04 |
|---|
| alcohol catabolic process | GO:0046164 |  | 3.404E-06 | 1.17E-03 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 1.386E-05 | 4.169E-03 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.386E-05 | 3.705E-03 |
|---|
| mRNA transcription | GO:0009299 |  | 1.386E-05 | 3.335E-03 |
|---|
| positive regulation of chemokine production | GO:0032722 |  | 1.386E-05 | 3.032E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 5.864E-08 | 2.094E-04 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.724E-07 | 3.079E-04 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 9.376E-06 | 0.0111604 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 1.104E-05 | 9.852E-03 |
|---|
| mRNA transcription | GO:0009299 |  | 1.104E-05 | 7.881E-03 |
|---|
| oxygen homeostasis | GO:0032364 |  | 1.104E-05 | 6.568E-03 |
|---|
| bHLH transcription factor binding | GO:0043425 |  | 1.104E-05 | 5.629E-03 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.104E-05 | 4.926E-03 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.104E-05 | 4.378E-03 |
|---|
| positive regulation of chemokine production | GO:0032722 |  | 1.104E-05 | 3.941E-03 |
|---|
| histone acetyltransferase binding | GO:0035035 |  | 1.104E-05 | 3.582E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.861E-08 | 1.044E-04 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.854E-07 | 3.382E-04 |
|---|
| response to light stimulus | GO:0009416 |  | 1.328E-06 | 1.615E-03 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 4.595E-06 | 4.19E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.191E-06 | 4.517E-03 |
|---|
| bHLH transcription factor binding | GO:0043425 |  | 6.839E-06 | 4.158E-03 |
|---|
| NF-kappaB-inducing kinase activity | GO:0004704 |  | 6.839E-06 | 3.564E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 6.839E-06 | 3.118E-03 |
|---|
| response to UV | GO:0009411 |  | 7.112E-06 | 2.883E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.025E-05 | 3.74E-03 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 1.025E-05 | 3.4E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.566E-08 | 3.767E-05 |
|---|
| glycolysis | GO:0006096 |  | 2.473E-07 | 2.975E-04 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.571E-07 | 2.062E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 5.613E-07 | 3.376E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.249E-06 | 6.008E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.074E-06 | 8.317E-04 |
|---|
| alcohol catabolic process | GO:0046164 |  | 3.404E-06 | 1.17E-03 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 1.386E-05 | 4.169E-03 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.386E-05 | 3.705E-03 |
|---|
| mRNA transcription | GO:0009299 |  | 1.386E-05 | 3.335E-03 |
|---|
| positive regulation of chemokine production | GO:0032722 |  | 1.386E-05 | 3.032E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 4.095E-08 | 9.418E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.424E-07 | 3.938E-04 |
|---|
| glycolysis | GO:0006096 |  | 7.916E-07 | 6.069E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.823E-06 | 1.048E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.621E-06 | 1.666E-03 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.494E-06 | 2.106E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 8.851E-06 | 2.908E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.61E-05 | 4.628E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.825E-05 | 4.665E-03 |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 2.649E-05 | 6.092E-03 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 2.649E-05 | 5.538E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 4.095E-08 | 9.418E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.424E-07 | 3.938E-04 |
|---|
| glycolysis | GO:0006096 |  | 7.916E-07 | 6.069E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.823E-06 | 1.048E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.621E-06 | 1.666E-03 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.494E-06 | 2.106E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 8.851E-06 | 2.908E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.61E-05 | 4.628E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.825E-05 | 4.665E-03 |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 2.649E-05 | 6.092E-03 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 2.649E-05 | 5.538E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 4.095E-08 | 9.418E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.424E-07 | 3.938E-04 |
|---|
| glycolysis | GO:0006096 |  | 7.916E-07 | 6.069E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.823E-06 | 1.048E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.621E-06 | 1.666E-03 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.494E-06 | 2.106E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 8.851E-06 | 2.908E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.61E-05 | 4.628E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.825E-05 | 4.665E-03 |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 2.649E-05 | 6.092E-03 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 2.649E-05 | 5.538E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 4.095E-08 | 9.418E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.424E-07 | 3.938E-04 |
|---|
| glycolysis | GO:0006096 |  | 7.916E-07 | 6.069E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.823E-06 | 1.048E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.621E-06 | 1.666E-03 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.494E-06 | 2.106E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 8.851E-06 | 2.908E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.61E-05 | 4.628E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.825E-05 | 4.665E-03 |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 2.649E-05 | 6.092E-03 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 2.649E-05 | 5.538E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.422E-08 | 8.12E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 4.085E-07 | 4.846E-04 |
|---|
| glycolysis | GO:0006096 |  | 9.742E-07 | 7.706E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.215E-06 | 1.314E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 4.121E-06 | 1.956E-03 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 6.036E-06 | 2.387E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 9.379E-06 | 3.18E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.706E-05 | 5.06E-03 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 2.345E-05 | 6.182E-03 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 2.345E-05 | 5.564E-03 |
|---|
| positive regulation of chemokine production | GO:0032722 |  | 2.345E-05 | 5.058E-03 |
|---|



