Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 3420-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1927 | 1.023e-02 | 1.223e-02 | 1.008e-01 |
|---|
| IPC-NIBC-129 | 0.2788 | 4.147e-02 | 5.216e-02 | 3.982e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2472 | 1.920e-04 | 4.400e-04 | 1.462e-02 |
|---|
| Loi_GPL570 | 0.4251 | 4.140e-02 | 4.108e-02 | 2.213e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1088 | 0.000e+00 | 0.000e+00 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2718 | 1.088e-01 | 1.092e-01 | 5.629e-01 |
|---|
| Parker_GPL887 | 0.4442 | 6.891e-02 | 8.580e-02 | 5.315e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.3627 | 9.028e-01 | 9.030e-01 | 9.999e-01 |
|---|
| Schmidt | 0.3307 | 3.184e-02 | 4.311e-02 | 3.013e-01 |
|---|
| Sotiriou | 0.2440 | 3.575e-03 | 1.136e-03 | 8.049e-02 |
|---|
| Wang | 0.1714 | 2.476e-01 | 1.983e-01 | 6.285e-01 |
|---|
| Zhang | 0.0264 | 1.184e-01 | 8.020e-02 | 2.787e-01 |
|---|
| Zhou | 0.3913 | 5.977e-02 | 6.306e-02 | 3.709e-01 |
|---|
Expression data for subnetwork 3420-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
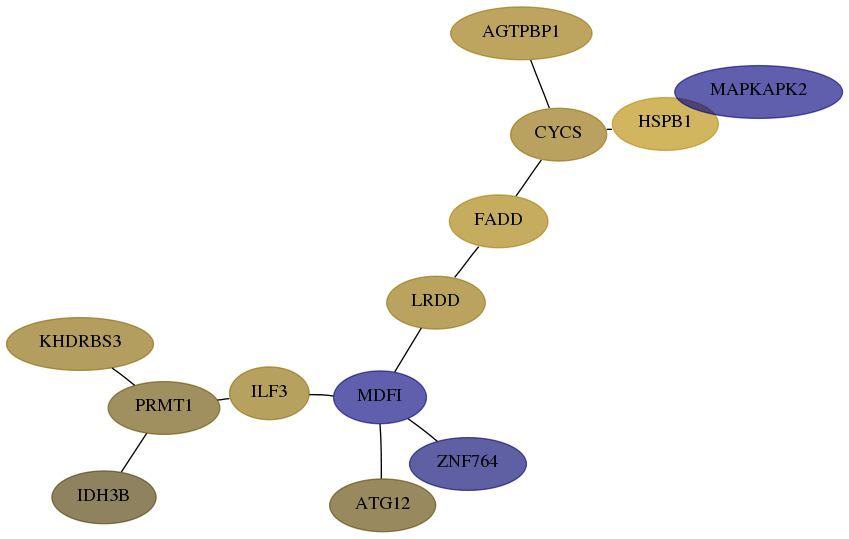
Score for each gene in subnetwork 3420-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| agtpbp1 |   | 11 | 21 | 17 | 13 | 0.086 | 0.219 | 0.009 | 0.181 | 0.071 | 0.078 | 0.296 | 0.026 | 0.110 | 0.151 | 0.103 | 0.064 | 0.226 |
|---|
| cycs |   | 10 | 25 | 17 | 14 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | -0.056 | 0.202 | 0.272 | 0.157 | 0.077 | 0.190 | 0.135 | 0.234 |
|---|
| znf764 |   | 1 | 164 | 64 | 87 | -0.034 | 0.055 | 0.106 | 0.123 | 0.058 | -0.153 | 0.156 | -0.074 | 0.043 | 0.092 | -0.003 | 0.139 | 0.052 |
|---|
| atg12 |   | 1 | 164 | 64 | 87 | 0.021 | -0.069 | 0.040 | 0.055 | 0.088 | 0.127 | -0.144 | -0.169 | 0.114 | 0.044 | 0.080 | 0.054 | 0.024 |
|---|
| ilf3 |   | 2 | 104 | 64 | 71 | 0.069 | 0.235 | 0.115 | 0.194 | 0.073 | 0.201 | 0.067 | 0.145 | 0.055 | 0.030 | -0.100 | 0.013 | 0.265 |
|---|
| fadd |   | 5 | 50 | 64 | 58 | 0.121 | -0.009 | 0.155 | 0.307 | 0.166 | 0.119 | 0.084 | 0.300 | 0.070 | 0.116 | 0.100 | 0.032 | 0.243 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| prmt1 |   | 2 | 104 | 64 | 71 | 0.029 | 0.172 | 0.254 | 0.122 | 0.144 | 0.192 | -0.083 | 0.169 | 0.003 | 0.154 | -0.065 | -0.147 | 0.219 |
|---|
| mapkapk2 |   | 13 | 19 | 17 | 12 | -0.050 | 0.102 | 0.156 | -0.116 | 0.090 | 0.168 | 0.303 | 0.074 | 0.171 | 0.117 | 0.085 | -0.124 | 0.312 |
|---|
| mdfi |   | 2 | 104 | 64 | 71 | -0.050 | 0.215 | 0.036 | -0.005 | 0.054 | 0.263 | 0.176 | 0.130 | 0.296 | -0.041 | 0.016 | 0.076 | 0.056 |
|---|
| khdrbs3 |   | 3 | 79 | 64 | 68 | 0.060 | 0.275 | -0.073 | 0.091 | 0.049 | 0.150 | 0.116 | 0.145 | 0.171 | 0.054 | -0.038 | 0.134 | 0.140 |
|---|
| idh3b |   | 1 | 164 | 64 | 87 | 0.015 | 0.065 | 0.079 | 0.275 | 0.100 | -0.143 | 0.526 | 0.130 | 0.114 | 0.016 | -0.027 | -0.019 | 0.180 |
|---|
| lrdd |   | 2 | 104 | 64 | 71 | 0.075 | 0.117 | -0.015 | 0.179 | 0.068 | 0.006 | 0.302 | 0.104 | 0.152 | 0.007 | -0.028 | 0.108 | 0.085 |
|---|
GO Enrichment output for subnetwork 3420-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| autophagic vacuole formation | GO:0000045 |  | 1.492E-08 | 3.431E-05 |
|---|
| macroautophagy | GO:0016236 |  | 2.981E-08 | 3.428E-05 |
|---|
| cellular response to starvation | GO:0009267 |  | 5.392E-07 | 4.134E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 1.205E-06 | 6.928E-04 |
|---|
| response to starvation | GO:0042594 |  | 1.43E-06 | 6.577E-04 |
|---|
| autophagy | GO:0006914 |  | 1.959E-06 | 7.511E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.959E-06 | 6.438E-04 |
|---|
| vesicle organization | GO:0016050 |  | 1.787E-05 | 5.139E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.056E-05 | 5.253E-03 |
|---|
| cytoplasmic sequestering of transcription factor | GO:0042994 |  | 4.923E-05 | 0.01132189 |
|---|
| cytoplasmic sequestering of protein | GO:0051220 |  | 6.149E-05 | 0.01285646 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.717E-08 | 4.195E-05 |
|---|
| autophagic vacuole formation | GO:0000045 |  | 3.676E-08 | 4.49E-05 |
|---|
| macroautophagy | GO:0016236 |  | 5.052E-08 | 4.114E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.076E-07 | 1.268E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 4.054E-07 | 1.981E-04 |
|---|
| cellular respiration | GO:0045333 |  | 4.264E-07 | 1.736E-04 |
|---|
| cellular response to starvation | GO:0009267 |  | 4.691E-07 | 1.637E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.691E-07 | 1.433E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 6.16E-07 | 1.672E-04 |
|---|
| response to starvation | GO:0042594 |  | 8.89E-07 | 2.172E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 8.89E-07 | 1.974E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.483E-08 | 5.975E-05 |
|---|
| autophagic vacuole formation | GO:0000045 |  | 3.723E-08 | 4.479E-05 |
|---|
| macroautophagy | GO:0016236 |  | 5.316E-08 | 4.263E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.001E-07 | 1.805E-04 |
|---|
| cellular response to starvation | GO:0009267 |  | 5.024E-07 | 2.417E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 5.858E-07 | 2.349E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 6.779E-07 | 2.33E-04 |
|---|
| cellular respiration | GO:0045333 |  | 6.914E-07 | 2.079E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 8.9E-07 | 2.379E-04 |
|---|
| response to starvation | GO:0042594 |  | 1.011E-06 | 2.432E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 1.011E-06 | 2.211E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.717E-08 | 4.195E-05 |
|---|
| autophagic vacuole formation | GO:0000045 |  | 3.676E-08 | 4.49E-05 |
|---|
| macroautophagy | GO:0016236 |  | 5.052E-08 | 4.114E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.076E-07 | 1.268E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 4.054E-07 | 1.981E-04 |
|---|
| cellular respiration | GO:0045333 |  | 4.264E-07 | 1.736E-04 |
|---|
| cellular response to starvation | GO:0009267 |  | 4.691E-07 | 1.637E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.691E-07 | 1.433E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 6.16E-07 | 1.672E-04 |
|---|
| response to starvation | GO:0042594 |  | 8.89E-07 | 2.172E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 8.89E-07 | 1.974E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.483E-08 | 5.975E-05 |
|---|
| autophagic vacuole formation | GO:0000045 |  | 3.723E-08 | 4.479E-05 |
|---|
| macroautophagy | GO:0016236 |  | 5.316E-08 | 4.263E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.001E-07 | 1.805E-04 |
|---|
| cellular response to starvation | GO:0009267 |  | 5.024E-07 | 2.417E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 5.858E-07 | 2.349E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 6.779E-07 | 2.33E-04 |
|---|
| cellular respiration | GO:0045333 |  | 6.914E-07 | 2.079E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 8.9E-07 | 2.379E-04 |
|---|
| response to starvation | GO:0042594 |  | 1.011E-06 | 2.432E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 1.011E-06 | 2.211E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| death receptor binding | GO:0005123 |  | 3.814E-08 | 1.362E-04 |
|---|
| tumor necrosis factor receptor superfamily binding | GO:0032813 |  | 2.459E-06 | 4.391E-03 |
|---|
| isocitrate dehydrogenase activity | GO:0004448 |  | 8.371E-06 | 9.964E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.756E-05 | 0.01567578 |
|---|
| autophagic vacuole formation | GO:0000045 |  | 1.756E-05 | 0.01254063 |
|---|
| macroautophagy | GO:0016236 |  | 2.34E-05 | 0.01392646 |
|---|
| cytoplasmic sequestering of transcription factor | GO:0042994 |  | 3.007E-05 | 0.0153392 |
|---|
| cytoplasmic sequestering of protein | GO:0051220 |  | 4.589E-05 | 0.02048326 |
|---|
| induction of apoptosis via death domain receptors | GO:0008625 |  | 5.504E-05 | 0.02183695 |
|---|
| negative regulation of transcription factor import into nucleus | GO:0042992 |  | 5.504E-05 | 0.01965325 |
|---|
| cellular respiration | GO:0045333 |  | 6.442E-05 | 0.02091404 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| death receptor binding | GO:0005123 |  | 2.861E-08 | 1.044E-04 |
|---|
| tumor necrosis factor receptor superfamily binding | GO:0032813 |  | 1.847E-06 | 3.368E-03 |
|---|
| isocitrate dehydrogenase activity | GO:0004448 |  | 6.839E-06 | 8.316E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.435E-05 | 0.0130844 |
|---|
| dorsal/ventral axis specification | GO:0009950 |  | 1.435E-05 | 0.01046752 |
|---|
| cerebellar Purkinje cell layer development | GO:0021680 |  | 1.435E-05 | 8.723E-03 |
|---|
| cerebellar cortex formation | GO:0021697 |  | 1.912E-05 | 9.964E-03 |
|---|
| cell differentiation in hindbrain | GO:0021533 |  | 2.457E-05 | 0.01120402 |
|---|
| cytoplasmic sequestering of transcription factor | GO:0042994 |  | 2.457E-05 | 9.959E-03 |
|---|
| autophagic vacuole formation | GO:0000045 |  | 3.07E-05 | 0.01119844 |
|---|
| cerebellar cortex morphogenesis | GO:0021696 |  | 3.75E-05 | 0.0124365 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.483E-08 | 5.975E-05 |
|---|
| autophagic vacuole formation | GO:0000045 |  | 3.723E-08 | 4.479E-05 |
|---|
| macroautophagy | GO:0016236 |  | 5.316E-08 | 4.263E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.001E-07 | 1.805E-04 |
|---|
| cellular response to starvation | GO:0009267 |  | 5.024E-07 | 2.417E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 5.858E-07 | 2.349E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 6.779E-07 | 2.33E-04 |
|---|
| cellular respiration | GO:0045333 |  | 6.914E-07 | 2.079E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 8.9E-07 | 2.379E-04 |
|---|
| response to starvation | GO:0042594 |  | 1.011E-06 | 2.432E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 1.011E-06 | 2.211E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| autophagic vacuole formation | GO:0000045 |  | 1.492E-08 | 3.431E-05 |
|---|
| macroautophagy | GO:0016236 |  | 2.981E-08 | 3.428E-05 |
|---|
| cellular response to starvation | GO:0009267 |  | 5.392E-07 | 4.134E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 1.205E-06 | 6.928E-04 |
|---|
| response to starvation | GO:0042594 |  | 1.43E-06 | 6.577E-04 |
|---|
| autophagy | GO:0006914 |  | 1.959E-06 | 7.511E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.959E-06 | 6.438E-04 |
|---|
| vesicle organization | GO:0016050 |  | 1.787E-05 | 5.139E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.056E-05 | 5.253E-03 |
|---|
| cytoplasmic sequestering of transcription factor | GO:0042994 |  | 4.923E-05 | 0.01132189 |
|---|
| cytoplasmic sequestering of protein | GO:0051220 |  | 6.149E-05 | 0.01285646 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| autophagic vacuole formation | GO:0000045 |  | 1.492E-08 | 3.431E-05 |
|---|
| macroautophagy | GO:0016236 |  | 2.981E-08 | 3.428E-05 |
|---|
| cellular response to starvation | GO:0009267 |  | 5.392E-07 | 4.134E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 1.205E-06 | 6.928E-04 |
|---|
| response to starvation | GO:0042594 |  | 1.43E-06 | 6.577E-04 |
|---|
| autophagy | GO:0006914 |  | 1.959E-06 | 7.511E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.959E-06 | 6.438E-04 |
|---|
| vesicle organization | GO:0016050 |  | 1.787E-05 | 5.139E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.056E-05 | 5.253E-03 |
|---|
| cytoplasmic sequestering of transcription factor | GO:0042994 |  | 4.923E-05 | 0.01132189 |
|---|
| cytoplasmic sequestering of protein | GO:0051220 |  | 6.149E-05 | 0.01285646 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| autophagic vacuole formation | GO:0000045 |  | 1.492E-08 | 3.431E-05 |
|---|
| macroautophagy | GO:0016236 |  | 2.981E-08 | 3.428E-05 |
|---|
| cellular response to starvation | GO:0009267 |  | 5.392E-07 | 4.134E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 1.205E-06 | 6.928E-04 |
|---|
| response to starvation | GO:0042594 |  | 1.43E-06 | 6.577E-04 |
|---|
| autophagy | GO:0006914 |  | 1.959E-06 | 7.511E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.959E-06 | 6.438E-04 |
|---|
| vesicle organization | GO:0016050 |  | 1.787E-05 | 5.139E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.056E-05 | 5.253E-03 |
|---|
| cytoplasmic sequestering of transcription factor | GO:0042994 |  | 4.923E-05 | 0.01132189 |
|---|
| cytoplasmic sequestering of protein | GO:0051220 |  | 6.149E-05 | 0.01285646 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| autophagic vacuole formation | GO:0000045 |  | 1.492E-08 | 3.431E-05 |
|---|
| macroautophagy | GO:0016236 |  | 2.981E-08 | 3.428E-05 |
|---|
| cellular response to starvation | GO:0009267 |  | 5.392E-07 | 4.134E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 1.205E-06 | 6.928E-04 |
|---|
| response to starvation | GO:0042594 |  | 1.43E-06 | 6.577E-04 |
|---|
| autophagy | GO:0006914 |  | 1.959E-06 | 7.511E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.959E-06 | 6.438E-04 |
|---|
| vesicle organization | GO:0016050 |  | 1.787E-05 | 5.139E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.056E-05 | 5.253E-03 |
|---|
| cytoplasmic sequestering of transcription factor | GO:0042994 |  | 4.923E-05 | 0.01132189 |
|---|
| cytoplasmic sequestering of protein | GO:0051220 |  | 6.149E-05 | 0.01285646 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| autophagic vacuole formation | GO:0000045 |  | 5.387E-08 | 1.278E-04 |
|---|
| macroautophagy | GO:0016236 |  | 8.612E-08 | 1.022E-04 |
|---|
| cellular response to starvation | GO:0009267 |  | 8.558E-07 | 6.769E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 1.737E-06 | 1.03E-03 |
|---|
| response to starvation | GO:0042594 |  | 2.024E-06 | 9.608E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 2.691E-06 | 1.064E-03 |
|---|
| autophagy | GO:0006914 |  | 4.431E-06 | 1.502E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.087E-05 | 6.192E-03 |
|---|
| vesicle organization | GO:0016050 |  | 2.577E-05 | 6.795E-03 |
|---|
| cytoplasmic sequestering of transcription factor | GO:0042994 |  | 4.998E-05 | 0.01186141 |
|---|
| cytoplasmic sequestering of protein | GO:0051220 |  | 6.244E-05 | 0.01346894 |
|---|



