Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 3251-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1961 | 1.065e-02 | 1.273e-02 | 1.039e-01 |
|---|
| IPC-NIBC-129 | 0.1985 | 2.850e-02 | 3.488e-02 | 3.014e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2452 | 5.775e-03 | 9.395e-03 | 1.872e-01 |
|---|
| Loi_GPL570 | 0.2686 | 1.229e-01 | 1.222e-01 | 4.717e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1555 | 9.000e-06 | 6.300e-05 | 7.200e-05 |
|---|
| Parker_GPL1390 | 0.2988 | 1.037e-01 | 1.037e-01 | 5.504e-01 |
|---|
| Parker_GPL887 | 0.2586 | 2.438e-01 | 2.758e-01 | 2.394e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3316 | 7.642e-02 | 5.352e-02 | 4.372e-01 |
|---|
| Schmidt | 0.2398 | 1.293e-02 | 1.826e-02 | 1.590e-01 |
|---|
| Sotiriou | 0.2995 | 3.002e-03 | 9.110e-04 | 7.108e-02 |
|---|
| Wang | 0.1841 | 1.190e-01 | 8.423e-02 | 4.087e-01 |
|---|
| Zhang | 0.2972 | 9.869e-02 | 6.447e-02 | 2.375e-01 |
|---|
| Zhou | 0.4019 | 8.029e-02 | 8.703e-02 | 4.707e-01 |
|---|
Expression data for subnetwork 3251-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
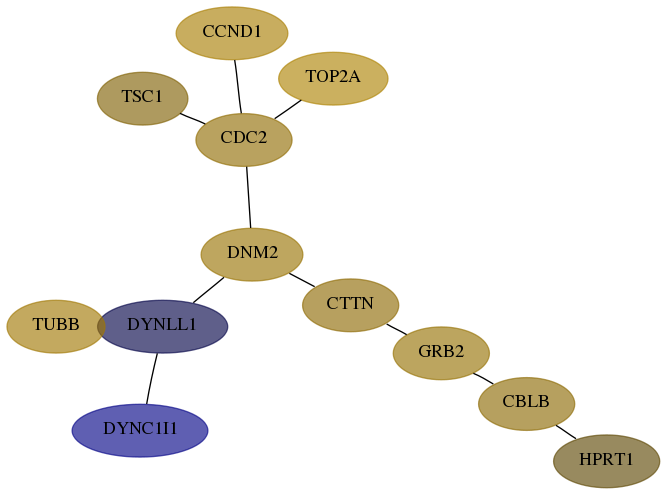
Score for each gene in subnetwork 3251-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| cttn |   | 9 | 27 | 44 | 37 | 0.067 | 0.065 | 0.123 | 0.238 | 0.126 | 0.263 | -0.172 | 0.216 | 0.069 | 0.115 | 0.129 | 0.025 | 0.279 |
|---|
| tubb |   | 8 | 33 | 93 | 80 | 0.107 | 0.221 | 0.070 | 0.090 | 0.095 | 0.278 | -0.325 | 0.185 | 0.139 | 0.202 | -0.061 | -0.142 | 0.101 |
|---|
| hprt1 |   | 1 | 164 | 93 | 111 | 0.022 | 0.159 | 0.088 | 0.172 | 0.150 | 0.302 | -0.126 | 0.203 | 0.112 | 0.106 | 0.137 | 0.179 | 0.207 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| dync1i1 |   | 2 | 104 | 93 | 97 | -0.058 | 0.102 | 0.055 | 0.124 | 0.054 | 0.147 | -0.008 | 0.056 | 0.082 | 0.036 | 0.119 | 0.166 | -0.245 |
|---|
| cblb |   | 2 | 104 | 93 | 97 | 0.066 | 0.051 | 0.077 | 0.055 | 0.058 | -0.023 | -0.391 | -0.166 | -0.005 | 0.045 | 0.089 | 0.041 | 0.101 |
|---|
| dnm2 |   | 6 | 43 | 93 | 81 | 0.089 | 0.127 | 0.114 | -0.049 | 0.065 | 0.251 | 0.291 | 0.177 | -0.039 | -0.048 | -0.099 | -0.044 | 0.262 |
|---|
| top2a |   | 11 | 21 | 93 | 76 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.072 | 0.234 | 0.273 |
|---|
| grb2 |   | 9 | 27 | 1 | 4 | 0.084 | -0.013 | 0.086 | -0.031 | 0.162 | -0.038 | 0.003 | 0.065 | 0.158 | 0.127 | -0.097 | 0.194 | -0.005 |
|---|
| dynll1 |   | 5 | 50 | 93 | 82 | -0.012 | 0.108 | 0.088 | 0.087 | 0.098 | 0.124 | 0.134 | 0.128 | 0.113 | 0.093 | 0.158 | 0.074 | 0.360 |
|---|
GO Enrichment output for subnetwork 3251-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 7.686E-12 | 1.768E-08 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 1.931E-10 | 2.22E-07 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 5.049E-10 | 3.871E-07 |
|---|
| cell killing | GO:0001906 |  | 1.528E-09 | 8.786E-07 |
|---|
| spindle organization | GO:0007051 |  | 4.131E-08 | 1.9E-05 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 8.203E-07 | 3.144E-04 |
|---|
| leukocyte mediated immunity | GO:0002443 |  | 1.419E-06 | 4.662E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.789E-06 | 8.018E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 3.505E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.371E-05 | 3.154E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.371E-05 | 2.867E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 1.539E-12 | 3.759E-09 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 6.445E-11 | 7.873E-08 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 1.518E-10 | 1.236E-07 |
|---|
| cell killing | GO:0001906 |  | 4.178E-10 | 2.552E-07 |
|---|
| spindle organization | GO:0007051 |  | 1.091E-08 | 5.329E-06 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 2.017E-07 | 8.213E-05 |
|---|
| leukocyte mediated immunity | GO:0002443 |  | 3.602E-07 | 1.257E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.975E-06 | 1.825E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 5.975E-06 | 1.622E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 6.935E-06 | 1.694E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.959E-06 | 1.99E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 2.18E-12 | 5.244E-09 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 9.128E-11 | 1.098E-07 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 2.149E-10 | 1.724E-07 |
|---|
| cell killing | GO:0001906 |  | 5.916E-10 | 3.559E-07 |
|---|
| spindle organization | GO:0007051 |  | 1.543E-08 | 7.426E-06 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 2.676E-07 | 1.073E-04 |
|---|
| leukocyte mediated immunity | GO:0002443 |  | 4.818E-07 | 1.656E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.155E-06 | 2.152E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 7.155E-06 | 1.913E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 7.155E-06 | 1.722E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.155E-06 | 1.565E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 1.539E-12 | 3.759E-09 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 6.445E-11 | 7.873E-08 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 1.518E-10 | 1.236E-07 |
|---|
| cell killing | GO:0001906 |  | 4.178E-10 | 2.552E-07 |
|---|
| spindle organization | GO:0007051 |  | 1.091E-08 | 5.329E-06 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 2.017E-07 | 8.213E-05 |
|---|
| leukocyte mediated immunity | GO:0002443 |  | 3.602E-07 | 1.257E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.975E-06 | 1.825E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 5.975E-06 | 1.622E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 6.935E-06 | 1.694E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 8.959E-06 | 1.99E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 2.18E-12 | 5.244E-09 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 9.128E-11 | 1.098E-07 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 2.149E-10 | 1.724E-07 |
|---|
| cell killing | GO:0001906 |  | 5.916E-10 | 3.559E-07 |
|---|
| spindle organization | GO:0007051 |  | 1.543E-08 | 7.426E-06 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 2.676E-07 | 1.073E-04 |
|---|
| leukocyte mediated immunity | GO:0002443 |  | 4.818E-07 | 1.656E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.155E-06 | 2.152E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 7.155E-06 | 1.913E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 7.155E-06 | 1.722E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.155E-06 | 1.565E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cytoplasmic dynein complex | GO:0005868 |  | 4.124E-09 | 1.473E-05 |
|---|
| dynein complex | GO:0030286 |  | 1.337E-06 | 2.387E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 4.009E-06 | 4.773E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.073E-06 | 5.421E-03 |
|---|
| spindle assembly | GO:0051225 |  | 6.073E-06 | 4.337E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.073E-06 | 3.614E-03 |
|---|
| DNA topoisomerase activity | GO:0003916 |  | 6.073E-06 | 3.098E-03 |
|---|
| nucleoside salvage | GO:0043174 |  | 6.073E-06 | 2.711E-03 |
|---|
| microtubule binding | GO:0008017 |  | 6.143E-06 | 2.438E-03 |
|---|
| lamellipodium | GO:0030027 |  | 7.446E-06 | 2.659E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.105E-06 | 2.956E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cytoplasmic dynein complex | GO:0005868 |  | 8.312E-09 | 3.032E-05 |
|---|
| dynein complex | GO:0030286 |  | 9.472E-07 | 1.728E-03 |
|---|
| lamellipodium | GO:0030027 |  | 4.356E-06 | 5.297E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 5.984E-06 | 5.458E-03 |
|---|
| grooming behavior | GO:0007625 |  | 5.984E-06 | 4.366E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 5.984E-06 | 3.639E-03 |
|---|
| purine salvage | GO:0043101 |  | 5.984E-06 | 3.119E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 5.984E-06 | 2.729E-03 |
|---|
| epidermal growth factor receptor binding | GO:0005154 |  | 5.984E-06 | 2.426E-03 |
|---|
| microtubule binding | GO:0008017 |  | 6.608E-06 | 2.41E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 6.608E-06 | 2.191E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 2.18E-12 | 5.244E-09 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 9.128E-11 | 1.098E-07 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 2.149E-10 | 1.724E-07 |
|---|
| cell killing | GO:0001906 |  | 5.916E-10 | 3.559E-07 |
|---|
| spindle organization | GO:0007051 |  | 1.543E-08 | 7.426E-06 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 2.676E-07 | 1.073E-04 |
|---|
| leukocyte mediated immunity | GO:0002443 |  | 4.818E-07 | 1.656E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.155E-06 | 2.152E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 7.155E-06 | 1.913E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 7.155E-06 | 1.722E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.155E-06 | 1.565E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 7.686E-12 | 1.768E-08 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 1.931E-10 | 2.22E-07 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 5.049E-10 | 3.871E-07 |
|---|
| cell killing | GO:0001906 |  | 1.528E-09 | 8.786E-07 |
|---|
| spindle organization | GO:0007051 |  | 4.131E-08 | 1.9E-05 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 8.203E-07 | 3.144E-04 |
|---|
| leukocyte mediated immunity | GO:0002443 |  | 1.419E-06 | 4.662E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.789E-06 | 8.018E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 3.505E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.371E-05 | 3.154E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.371E-05 | 2.867E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 7.686E-12 | 1.768E-08 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 1.931E-10 | 2.22E-07 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 5.049E-10 | 3.871E-07 |
|---|
| cell killing | GO:0001906 |  | 1.528E-09 | 8.786E-07 |
|---|
| spindle organization | GO:0007051 |  | 4.131E-08 | 1.9E-05 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 8.203E-07 | 3.144E-04 |
|---|
| leukocyte mediated immunity | GO:0002443 |  | 1.419E-06 | 4.662E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.789E-06 | 8.018E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 3.505E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.371E-05 | 3.154E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.371E-05 | 2.867E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 7.686E-12 | 1.768E-08 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 1.931E-10 | 2.22E-07 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 5.049E-10 | 3.871E-07 |
|---|
| cell killing | GO:0001906 |  | 1.528E-09 | 8.786E-07 |
|---|
| spindle organization | GO:0007051 |  | 4.131E-08 | 1.9E-05 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 8.203E-07 | 3.144E-04 |
|---|
| leukocyte mediated immunity | GO:0002443 |  | 1.419E-06 | 4.662E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.789E-06 | 8.018E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 3.505E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.371E-05 | 3.154E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.371E-05 | 2.867E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 7.686E-12 | 1.768E-08 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 1.931E-10 | 2.22E-07 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 5.049E-10 | 3.871E-07 |
|---|
| cell killing | GO:0001906 |  | 1.528E-09 | 8.786E-07 |
|---|
| spindle organization | GO:0007051 |  | 4.131E-08 | 1.9E-05 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 8.203E-07 | 3.144E-04 |
|---|
| leukocyte mediated immunity | GO:0002443 |  | 1.419E-06 | 4.662E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.789E-06 | 8.018E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 3.505E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.371E-05 | 3.154E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.371E-05 | 2.867E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 1.681E-11 | 3.99E-08 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 1.409E-10 | 1.672E-07 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 3.686E-10 | 2.915E-07 |
|---|
| cell killing | GO:0001906 |  | 1.116E-09 | 6.618E-07 |
|---|
| spindle organization | GO:0007051 |  | 4.498E-08 | 2.135E-05 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 7.735E-07 | 3.059E-04 |
|---|
| leukocyte mediated immunity | GO:0002443 |  | 1.296E-06 | 4.393E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.695E-06 | 7.994E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.166E-05 | 3.074E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.166E-05 | 2.766E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.166E-05 | 2.515E-03 |
|---|



