Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 29110-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2342 | 6.154e-03 | 7.328e-03 | 6.851e-02 |
|---|
| IPC-NIBC-129 | 0.2282 | 6.158e-02 | 7.936e-02 | 5.165e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2718 | 1.407e-02 | 2.093e-02 | 3.212e-01 |
|---|
| Loi_GPL570 | 0.2269 | 8.262e-02 | 8.210e-02 | 3.641e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1855 | 1.240e-02 | 3.189e-02 | 3.733e-01 |
|---|
| Parker_GPL1390 | 0.2431 | 6.074e-02 | 5.784e-02 | 4.193e-01 |
|---|
| Parker_GPL887 | 0.1031 | 6.381e-01 | 6.669e-01 | 6.896e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3194 | 1.441e-01 | 1.117e-01 | 6.369e-01 |
|---|
| Schmidt | 0.1304 | 9.941e-03 | 1.421e-02 | 1.302e-01 |
|---|
| Sotiriou | 0.3155 | 1.158e-03 | 2.730e-04 | 3.533e-02 |
|---|
| Wang | 0.1686 | 7.340e-02 | 4.793e-02 | 2.975e-01 |
|---|
| Zhang | 0.2353 | 1.232e-01 | 8.408e-02 | 2.884e-01 |
|---|
| Zhou | 0.4590 | 9.001e-02 | 9.853e-02 | 5.123e-01 |
|---|
Expression data for subnetwork 29110-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
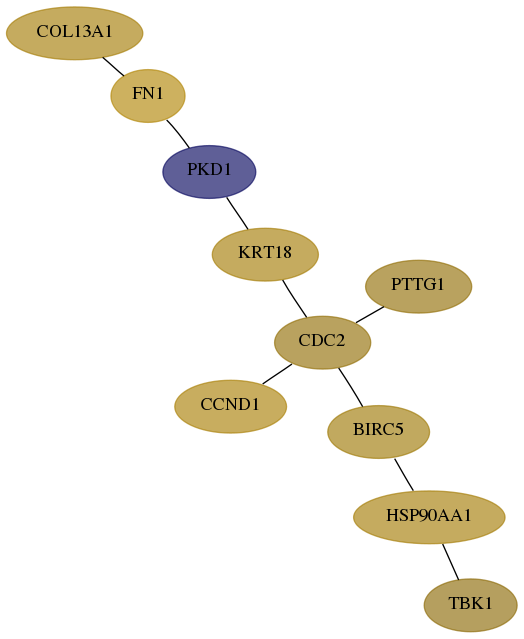
Score for each gene in subnetwork 29110-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| pttg1 |   | 10 | 25 | 31 | 21 | 0.072 | 0.270 | 0.175 | 0.183 | 0.200 | 0.208 | 0.347 | 0.334 | 0.224 | 0.204 | 0.147 | 0.210 | 0.309 |
|---|
| krt18 |   | 15 | 15 | 230 | 195 | 0.112 | -0.078 | 0.147 | 0.150 | 0.161 | -0.086 | -0.204 | 0.083 | -0.099 | 0.118 | 0.086 | -0.114 | 0.222 |
|---|
| col13a1 |   | 1 | 164 | 350 | 350 | 0.117 | 0.153 | 0.132 | 0.044 | 0.112 | 0.182 | -0.078 | 0.098 | -0.050 | 0.099 | 0.068 | 0.165 | 0.135 |
|---|
| hsp90aa1 |   | 11 | 21 | 234 | 204 | 0.116 | 0.030 | 0.164 | 0.076 | 0.119 | 0.155 | -0.199 | 0.169 | 0.069 | 0.102 | 0.052 | -0.011 | 0.399 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| tbk1 |   | 1 | 164 | 350 | 350 | 0.064 | 0.054 | 0.027 | -0.010 | 0.091 | 0.008 | -0.142 | -0.124 | -0.256 | 0.002 | 0.115 | 0.089 | 0.255 |
|---|
| pkd1 |   | 9 | 27 | 234 | 205 | -0.021 | 0.055 | 0.007 | 0.037 | 0.071 | 0.184 | 0.256 | -0.217 | -0.068 | -0.001 | 0.046 | 0.077 | 0.151 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| fn1 |   | 3 | 79 | 234 | 222 | 0.156 | 0.240 | -0.023 | -0.035 | 0.017 | 0.089 | -0.120 | 0.077 | -0.045 | -0.063 | 0.109 | 0.067 | 0.230 |
|---|
| birc5 |   | 14 | 17 | 162 | 140 | 0.101 | 0.216 | 0.190 | 0.134 | 0.223 | 0.149 | 0.166 | 0.202 | 0.236 | 0.241 | 0.124 | 0.205 | 0.400 |
|---|
GO Enrichment output for subnetwork 29110-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| endochondral ossification | GO:0001958 |  | 1.652E-05 | 0.03799068 |
|---|
| protein refolding | GO:0042026 |  | 2.476E-05 | 0.0284702 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.476E-05 | 0.01898013 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.476E-05 | 0.0142351 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.723E-05 | 0.01252434 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 0.01327545 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 3.463E-05 | 0.01137896 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 3.463E-05 | 9.957E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 4.464E-05 | 0.01140819 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.614E-05 | 0.01061186 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 5.927E-05 | 0.01239354 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| endochondral ossification | GO:0001958 |  | 4.743E-06 | 0.01158756 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.206E-06 | 6.359E-03 |
|---|
| protein refolding | GO:0042026 |  | 7.112E-06 | 5.791E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 9.952E-06 | 6.078E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 9.952E-06 | 4.863E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 9.952E-06 | 4.052E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 9.952E-06 | 3.473E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 1.187E-05 | 3.625E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.326E-05 | 3.6E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.326E-05 | 3.24E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.326E-05 | 2.946E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| endochondral ossification | GO:0001958 |  | 6.077E-06 | 0.01462147 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.511E-06 | 9.036E-03 |
|---|
| protein refolding | GO:0042026 |  | 9.111E-06 | 7.307E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 1.275E-05 | 7.669E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 1.275E-05 | 6.135E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.275E-05 | 5.113E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.275E-05 | 4.382E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 1.404E-05 | 4.221E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.699E-05 | 4.542E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.699E-05 | 4.088E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.699E-05 | 3.716E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| endochondral ossification | GO:0001958 |  | 4.743E-06 | 0.01158756 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.206E-06 | 6.359E-03 |
|---|
| protein refolding | GO:0042026 |  | 7.112E-06 | 5.791E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 9.952E-06 | 6.078E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 9.952E-06 | 4.863E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 9.952E-06 | 4.052E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 9.952E-06 | 3.473E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 1.187E-05 | 3.625E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.326E-05 | 3.6E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.326E-05 | 3.24E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.326E-05 | 2.946E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| endochondral ossification | GO:0001958 |  | 6.077E-06 | 0.01462147 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.511E-06 | 9.036E-03 |
|---|
| protein refolding | GO:0042026 |  | 9.111E-06 | 7.307E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 1.275E-05 | 7.669E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 1.275E-05 | 6.135E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.275E-05 | 5.113E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.275E-05 | 4.382E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 1.404E-05 | 4.221E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.699E-05 | 4.542E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.699E-05 | 4.088E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.699E-05 | 3.716E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cysteine-type endopeptidase inhibitor activity | GO:0004869 |  | 9.053E-07 | 3.233E-03 |
|---|
| regulation of Rab protein signal transduction | GO:0032483 |  | 2.195E-06 | 3.92E-03 |
|---|
| Rab GTPase activator activity | GO:0005097 |  | 2.368E-06 | 2.818E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.142E-06 | 3.697E-03 |
|---|
| nitric-oxide synthase regulator activity | GO:0030235 |  | 4.142E-06 | 2.958E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.21E-06 | 3.696E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 6.21E-06 | 3.168E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 6.462E-06 | 2.884E-03 |
|---|
| centriolar satellite | GO:0034451 |  | 8.691E-06 | 3.448E-03 |
|---|
| fibrinogen complex | GO:0005577 |  | 8.691E-06 | 3.104E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 9.619E-06 | 3.123E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| midbody | GO:0030496 |  | 2.28E-07 | 8.318E-04 |
|---|
| spindle microtubule | GO:0005876 |  | 8.423E-07 | 1.536E-03 |
|---|
| microtubule organizing center part | GO:0044450 |  | 5.071E-06 | 6.167E-03 |
|---|
| cysteine-type endopeptidase inhibitor activity | GO:0004869 |  | 5.574E-06 | 5.084E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.082E-05 | 7.897E-03 |
|---|
| nitric-oxide synthase regulator activity | GO:0030235 |  | 1.082E-05 | 6.581E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 1.082E-05 | 5.64E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 1.082E-05 | 4.935E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.252E-05 | 5.076E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.622E-05 | 5.919E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.622E-05 | 5.381E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| endochondral ossification | GO:0001958 |  | 6.077E-06 | 0.01462147 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.511E-06 | 9.036E-03 |
|---|
| protein refolding | GO:0042026 |  | 9.111E-06 | 7.307E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 1.275E-05 | 7.669E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 1.275E-05 | 6.135E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.275E-05 | 5.113E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.275E-05 | 4.382E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 1.404E-05 | 4.221E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.699E-05 | 4.542E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.699E-05 | 4.088E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.699E-05 | 3.716E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| endochondral ossification | GO:0001958 |  | 1.652E-05 | 0.03799068 |
|---|
| protein refolding | GO:0042026 |  | 2.476E-05 | 0.0284702 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.476E-05 | 0.01898013 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.476E-05 | 0.0142351 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.723E-05 | 0.01252434 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 0.01327545 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 3.463E-05 | 0.01137896 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 3.463E-05 | 9.957E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 4.464E-05 | 0.01140819 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.614E-05 | 0.01061186 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 5.927E-05 | 0.01239354 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| endochondral ossification | GO:0001958 |  | 1.652E-05 | 0.03799068 |
|---|
| protein refolding | GO:0042026 |  | 2.476E-05 | 0.0284702 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.476E-05 | 0.01898013 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.476E-05 | 0.0142351 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.723E-05 | 0.01252434 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 0.01327545 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 3.463E-05 | 0.01137896 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 3.463E-05 | 9.957E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 4.464E-05 | 0.01140819 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.614E-05 | 0.01061186 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 5.927E-05 | 0.01239354 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| endochondral ossification | GO:0001958 |  | 1.652E-05 | 0.03799068 |
|---|
| protein refolding | GO:0042026 |  | 2.476E-05 | 0.0284702 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.476E-05 | 0.01898013 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.476E-05 | 0.0142351 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.723E-05 | 0.01252434 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 0.01327545 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 3.463E-05 | 0.01137896 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 3.463E-05 | 9.957E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 4.464E-05 | 0.01140819 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.614E-05 | 0.01061186 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 5.927E-05 | 0.01239354 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| endochondral ossification | GO:0001958 |  | 1.652E-05 | 0.03799068 |
|---|
| protein refolding | GO:0042026 |  | 2.476E-05 | 0.0284702 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.476E-05 | 0.01898013 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.476E-05 | 0.0142351 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.723E-05 | 0.01252434 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 0.01327545 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 3.463E-05 | 0.01137896 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 3.463E-05 | 9.957E-03 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 4.464E-05 | 0.01140819 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.614E-05 | 0.01061186 |
|---|
| regulation of exit from mitosis | GO:0007096 |  | 5.927E-05 | 0.01239354 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| detection of mechanical stimulus | GO:0050982 |  | 1.91E-09 | 4.531E-06 |
|---|
| cartilage condensation | GO:0001502 |  | 2.671E-09 | 3.169E-06 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 1.484E-08 | 1.174E-05 |
|---|
| cell-matrix adhesion | GO:0007160 |  | 3.816E-08 | 2.264E-05 |
|---|
| protein refolding | GO:0042026 |  | 4.451E-08 | 2.112E-05 |
|---|
| cell-substrate adhesion | GO:0031589 |  | 5.39E-08 | 2.132E-05 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 7.782E-08 | 2.638E-05 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 1.215E-07 | 3.603E-05 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.244E-07 | 3.28E-05 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 3.523E-07 | 8.36E-05 |
|---|
| detection of abiotic stimulus | GO:0009582 |  | 5.031E-07 | 1.085E-04 |
|---|



