Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 291-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2080 | 4.938e-03 | 5.868e-03 | 5.775e-02 |
|---|
| IPC-NIBC-129 | 0.2963 | 3.046e-02 | 3.746e-02 | 3.173e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2824 | 5.300e-04 | 1.098e-03 | 3.329e-02 |
|---|
| Loi_GPL570 | 0.3787 | 3.247e-02 | 3.220e-02 | 1.837e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1383 | 3.000e-06 | 2.200e-05 | 1.100e-05 |
|---|
| Parker_GPL1390 | 0.2941 | 8.782e-02 | 8.654e-02 | 5.075e-01 |
|---|
| Parker_GPL887 | 0.3230 | 1.586e-01 | 1.855e-01 | 1.447e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3810 | 4.063e-01 | 3.690e-01 | 9.348e-01 |
|---|
| Schmidt | 0.2566 | 7.614e-03 | 1.100e-02 | 1.056e-01 |
|---|
| Sotiriou | 0.3318 | 2.814e-03 | 8.400e-04 | 6.787e-02 |
|---|
| Wang | 0.2019 | 1.564e-01 | 1.159e-01 | 4.839e-01 |
|---|
| Zhang | 0.1280 | 7.642e-02 | 4.747e-02 | 1.887e-01 |
|---|
| Zhou | 0.4057 | 5.015e-02 | 5.202e-02 | 3.180e-01 |
|---|
Expression data for subnetwork 291-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
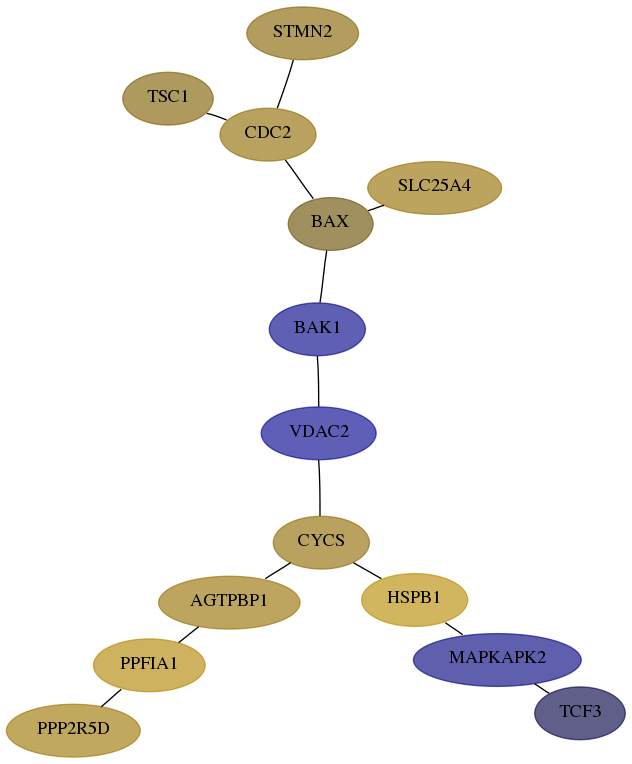
Score for each gene in subnetwork 291-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| ppp2r5d |   | 3 | 79 | 17 | 22 | 0.098 | 0.040 | 0.057 | 0.270 | 0.092 | 0.237 | 0.299 | 0.241 | 0.017 | 0.103 | 0.033 | 0.048 | 0.162 |
|---|
| agtpbp1 |   | 11 | 21 | 17 | 13 | 0.086 | 0.219 | 0.009 | 0.181 | 0.071 | 0.078 | 0.296 | 0.026 | 0.110 | 0.151 | 0.103 | 0.064 | 0.226 |
|---|
| stmn2 |   | 29 | 8 | 1 | 3 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.075 | 0.113 | -0.039 |
|---|
| cycs |   | 10 | 25 | 17 | 14 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | -0.056 | 0.202 | 0.272 | 0.157 | 0.077 | 0.190 | 0.135 | 0.234 |
|---|
| slc25a4 |   | 3 | 79 | 17 | 22 | 0.084 | 0.041 | 0.064 | 0.086 | 0.187 | 0.117 | 0.044 | 0.166 | 0.004 | 0.178 | 0.108 | 0.025 | 0.188 |
|---|
| bax |   | 9 | 27 | 17 | 15 | 0.029 | 0.185 | 0.233 | 0.192 | 0.088 | -0.043 | 0.184 | 0.172 | 0.175 | 0.019 | -0.094 | 0.041 | 0.235 |
|---|
| ppfia1 |   | 8 | 33 | 17 | 16 | 0.164 | 0.080 | 0.149 | 0.269 | 0.151 | 0.214 | 0.093 | 0.225 | 0.036 | 0.176 | 0.153 | 0.127 | 0.272 |
|---|
| tcf3 |   | 3 | 79 | 17 | 22 | -0.012 | 0.282 | 0.055 | 0.045 | 0.037 | 0.176 | 0.318 | 0.006 | 0.094 | -0.056 | 0.040 | 0.001 | 0.167 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| vdac2 |   | 1 | 164 | 17 | 42 | -0.071 | 0.176 | 0.131 | 0.212 | 0.187 | 0.208 | 0.007 | 0.086 | 0.035 | 0.069 | 0.037 | 0.048 | 0.248 |
|---|
| bak1 |   | 1 | 164 | 17 | 42 | -0.059 | 0.116 | 0.129 | 0.159 | 0.079 | -0.007 | 0.382 | 0.185 | 0.079 | 0.038 | -0.048 | 0.019 | undef |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| mapkapk2 |   | 13 | 19 | 17 | 12 | -0.050 | 0.102 | 0.156 | -0.116 | 0.090 | 0.168 | 0.303 | 0.074 | 0.171 | 0.117 | 0.085 | -0.124 | 0.312 |
|---|
GO Enrichment output for subnetwork 291-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.181E-08 | 1.882E-04 |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.43E-07 | 1.645E-04 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 1.43E-07 | 1.096E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 7.205E-07 | 4.143E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.13E-06 | 5.196E-04 |
|---|
| regulation of protein binding | GO:0043393 |  | 2.265E-06 | 8.683E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.854E-06 | 1.266E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.354E-06 | 1.252E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 5.81E-06 | 1.485E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.649E-05 | 6.092E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.649E-05 | 5.538E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 4.297E-11 | 1.05E-07 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.033E-09 | 1.262E-06 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.032E-09 | 1.655E-06 |
|---|
| regulation of caspase activity | GO:0043281 |  | 7.731E-09 | 4.721E-06 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 9.463E-09 | 4.624E-06 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 1.303E-08 | 5.304E-06 |
|---|
| B cell lineage commitment | GO:0002326 |  | 2.545E-08 | 8.883E-06 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 2.545E-08 | 7.773E-06 |
|---|
| regulation of protein binding | GO:0043393 |  | 4.914E-07 | 1.334E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 4.914E-07 | 1.2E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 9.587E-07 | 2.129E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.332E-11 | 2.005E-07 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.904E-09 | 2.291E-06 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 3.848E-09 | 3.086E-06 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.527E-08 | 9.187E-06 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.756E-08 | 8.449E-06 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 2.447E-08 | 9.812E-06 |
|---|
| B cell lineage commitment | GO:0002326 |  | 4.182E-08 | 1.437E-05 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.182E-08 | 1.258E-05 |
|---|
| regulation of protein binding | GO:0043393 |  | 8.065E-07 | 2.156E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 8.065E-07 | 1.94E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.573E-06 | 3.44E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 4.297E-11 | 1.05E-07 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.033E-09 | 1.262E-06 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.032E-09 | 1.655E-06 |
|---|
| regulation of caspase activity | GO:0043281 |  | 7.731E-09 | 4.721E-06 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 9.463E-09 | 4.624E-06 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 1.303E-08 | 5.304E-06 |
|---|
| B cell lineage commitment | GO:0002326 |  | 2.545E-08 | 8.883E-06 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 2.545E-08 | 7.773E-06 |
|---|
| regulation of protein binding | GO:0043393 |  | 4.914E-07 | 1.334E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 4.914E-07 | 1.2E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 9.587E-07 | 2.129E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.332E-11 | 2.005E-07 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.904E-09 | 2.291E-06 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 3.848E-09 | 3.086E-06 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.527E-08 | 9.187E-06 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.756E-08 | 8.449E-06 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 2.447E-08 | 9.812E-06 |
|---|
| B cell lineage commitment | GO:0002326 |  | 4.182E-08 | 1.437E-05 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.182E-08 | 1.258E-05 |
|---|
| regulation of protein binding | GO:0043393 |  | 8.065E-07 | 2.156E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 8.065E-07 | 1.94E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.573E-06 | 3.44E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.874E-08 | 6.693E-05 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 1.874E-08 | 3.346E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 5.823E-08 | 6.932E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 9.633E-08 | 8.599E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.707E-07 | 1.933E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 3.05E-07 | 1.816E-04 |
|---|
| mitochondrial outer membrane | GO:0005741 |  | 3.425E-07 | 1.747E-04 |
|---|
| regulation of protein binding | GO:0043393 |  | 3.623E-07 | 1.617E-04 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 4.05E-07 | 1.607E-04 |
|---|
| protein phosphatase type 2A complex | GO:0000159 |  | 4.345E-07 | 1.552E-04 |
|---|
| organelle outer membrane | GO:0031968 |  | 6.44E-07 | 2.091E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 1.242E-08 | 4.531E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.172E-08 | 3.962E-05 |
|---|
| regulation of mitochondrial membrane potential | GO:0051881 |  | 7.435E-08 | 9.041E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.25E-07 | 2.052E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.458E-07 | 2.523E-04 |
|---|
| protein phosphatase type 2A complex | GO:0000159 |  | 4.197E-07 | 2.552E-04 |
|---|
| mitochondrial outer membrane | GO:0005741 |  | 4.355E-07 | 2.27E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 7.024E-07 | 3.203E-04 |
|---|
| mitochondrion organization | GO:0007005 |  | 7.792E-07 | 3.158E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 8.19E-07 | 2.988E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 8.19E-07 | 2.716E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.332E-11 | 2.005E-07 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.904E-09 | 2.291E-06 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 3.848E-09 | 3.086E-06 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.527E-08 | 9.187E-06 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.756E-08 | 8.449E-06 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 2.447E-08 | 9.812E-06 |
|---|
| B cell lineage commitment | GO:0002326 |  | 4.182E-08 | 1.437E-05 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.182E-08 | 1.258E-05 |
|---|
| regulation of protein binding | GO:0043393 |  | 8.065E-07 | 2.156E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 8.065E-07 | 1.94E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.573E-06 | 3.44E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.181E-08 | 1.882E-04 |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.43E-07 | 1.645E-04 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 1.43E-07 | 1.096E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 7.205E-07 | 4.143E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.13E-06 | 5.196E-04 |
|---|
| regulation of protein binding | GO:0043393 |  | 2.265E-06 | 8.683E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.854E-06 | 1.266E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.354E-06 | 1.252E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 5.81E-06 | 1.485E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.649E-05 | 6.092E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.649E-05 | 5.538E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.181E-08 | 1.882E-04 |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.43E-07 | 1.645E-04 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 1.43E-07 | 1.096E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 7.205E-07 | 4.143E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.13E-06 | 5.196E-04 |
|---|
| regulation of protein binding | GO:0043393 |  | 2.265E-06 | 8.683E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.854E-06 | 1.266E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.354E-06 | 1.252E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 5.81E-06 | 1.485E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.649E-05 | 6.092E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.649E-05 | 5.538E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.181E-08 | 1.882E-04 |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.43E-07 | 1.645E-04 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 1.43E-07 | 1.096E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 7.205E-07 | 4.143E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.13E-06 | 5.196E-04 |
|---|
| regulation of protein binding | GO:0043393 |  | 2.265E-06 | 8.683E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.854E-06 | 1.266E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.354E-06 | 1.252E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 5.81E-06 | 1.485E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.649E-05 | 6.092E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.649E-05 | 5.538E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.181E-08 | 1.882E-04 |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.43E-07 | 1.645E-04 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 1.43E-07 | 1.096E-04 |
|---|
| activation of caspase activity | GO:0006919 |  | 7.205E-07 | 4.143E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.13E-06 | 5.196E-04 |
|---|
| regulation of protein binding | GO:0043393 |  | 2.265E-06 | 8.683E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.854E-06 | 1.266E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.354E-06 | 1.252E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 5.81E-06 | 1.485E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.649E-05 | 6.092E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.649E-05 | 5.538E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 9.718E-08 | 2.306E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.047E-05 | 0.02429314 |
|---|
| B cell differentiation | GO:0030183 |  | 2.656E-05 | 0.02100812 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.068E-05 | 0.01820337 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 3.068E-05 | 0.01456269 |
|---|
| activation of caspase activity | GO:0006919 |  | 4.057E-05 | 0.0160442 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.292E-05 | 0.01454951 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.538E-05 | 0.01642724 |
|---|
| neurotransmitter uptake | GO:0001504 |  | 5.717E-05 | 0.01507473 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 5.717E-05 | 0.01356726 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 9.172E-05 | 0.01978645 |
|---|



