Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1537-1-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2084 | 8.712e-03 | 1.040e-02 | 8.932e-02 |
|---|
| IPC-NIBC-129 | 0.1982 | 3.790e-02 | 4.737e-02 | 3.734e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2550 | 4.131e-03 | 6.951e-03 | 1.502e-01 |
|---|
| Loi_GPL570 | 0.2842 | 1.232e-01 | 1.226e-01 | 4.726e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1511 | 1.000e-06 | 1.200e-05 | 4.000e-06 |
|---|
| Parker_GPL1390 | 0.2783 | 8.739e-02 | 8.608e-02 | 5.063e-01 |
|---|
| Parker_GPL887 | 0.5364 | 3.599e-02 | 4.700e-02 | 2.402e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.4093 | 9.306e-01 | 9.322e-01 | 1.000e+00 |
|---|
| Schmidt | 0.2658 | 5.977e-03 | 8.728e-03 | 8.698e-02 |
|---|
| Sotiriou | 0.3465 | 8.038e-03 | 3.153e-03 | 1.405e-01 |
|---|
| Wang | 0.2302 | 1.277e-01 | 9.151e-02 | 4.274e-01 |
|---|
| Zhang | 0.0207 | 5.060e-02 | 2.897e-02 | 1.286e-01 |
|---|
| Zhou | 0.3421 | 3.818e-02 | 3.850e-02 | 2.461e-01 |
|---|
Expression data for subnetwork 1537-1-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
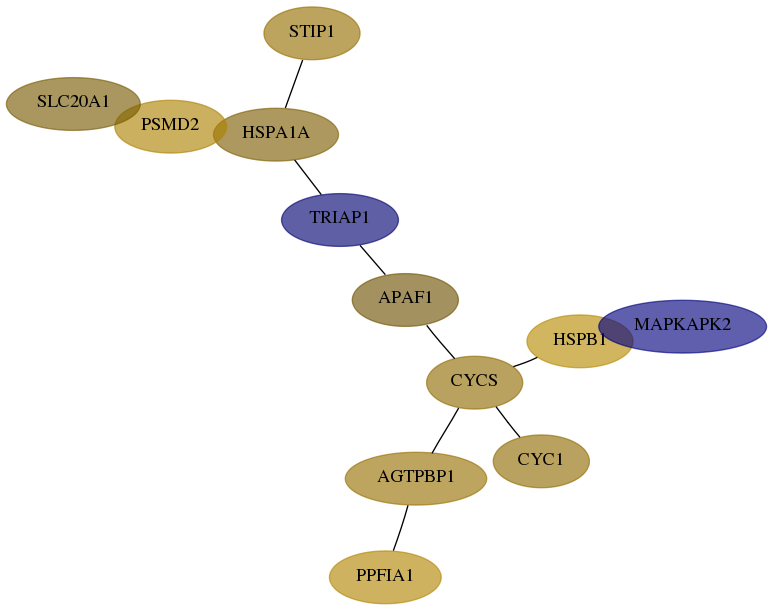
Score for each gene in subnetwork 1537-1-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| agtpbp1 |   | 11 | 21 | 17 | 13 | 0.086 | 0.219 | 0.009 | 0.181 | 0.071 | 0.078 | 0.296 | 0.026 | 0.110 | 0.151 | 0.103 | 0.064 | 0.226 |
|---|
| psmd2 |   | 7 | 38 | 37 | 28 | 0.147 | 0.322 | 0.122 | 0.146 | 0.143 | 0.246 | 0.444 | 0.194 | 0.318 | 0.151 | 0.154 | 0.042 | 0.207 |
|---|
| cycs |   | 10 | 25 | 17 | 14 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | -0.056 | 0.202 | 0.272 | 0.157 | 0.077 | 0.190 | 0.135 | 0.234 |
|---|
| ppfia1 |   | 8 | 33 | 17 | 16 | 0.164 | 0.080 | 0.149 | 0.269 | 0.151 | 0.214 | 0.093 | 0.225 | 0.036 | 0.176 | 0.153 | 0.127 | 0.272 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| triap1 |   | 1 | 164 | 37 | 65 | -0.037 | 0.067 | 0.191 | 0.117 | 0.140 | 0.281 | -0.288 | 0.123 | 0.163 | 0.130 | 0.142 | 0.078 | 0.249 |
|---|
| cyc1 |   | 1 | 164 | 37 | 65 | 0.072 | 0.077 | 0.066 | 0.193 | 0.165 | -0.125 | 0.569 | 0.253 | 0.104 | 0.189 | 0.082 | 0.064 | 0.140 |
|---|
| mapkapk2 |   | 13 | 19 | 17 | 12 | -0.050 | 0.102 | 0.156 | -0.116 | 0.090 | 0.168 | 0.303 | 0.074 | 0.171 | 0.117 | 0.085 | -0.124 | 0.312 |
|---|
| stip1 |   | 1 | 164 | 37 | 65 | 0.082 | 0.158 | 0.188 | 0.072 | 0.192 | 0.093 | 0.211 | 0.178 | 0.134 | 0.203 | 0.001 | 0.044 | 0.138 |
|---|
| slc20a1 |   | 7 | 38 | 37 | 28 | 0.041 | 0.039 | 0.109 | 0.170 | 0.101 | 0.121 | 0.326 | 0.086 | 0.048 | 0.124 | 0.121 | -0.050 | 0.299 |
|---|
| apaf1 |   | 2 | 104 | 37 | 46 | 0.033 | 0.135 | 0.021 | -0.049 | 0.072 | 0.087 | 0.202 | 0.001 | 0.120 | 0.008 | 0.026 | -0.004 | 0.228 |
|---|
| hspa1a |   | 4 | 60 | 37 | 39 | 0.047 | -0.046 | 0.070 | 0.019 | 0.160 | 0.104 | 0.230 | 0.108 | 0.121 | 0.110 | 0.175 | -0.055 | 0.192 |
|---|
GO Enrichment output for subnetwork 1537-1-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.981E-08 | 6.856E-05 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in cell cycle arrest | GO:0006977 |  | 1.371E-05 | 0.0157705 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.787E-05 | 0.01370377 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.495E-05 | 0.01434893 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 2.876E-05 | 0.01322803 |
|---|
| phosphate transport | GO:0006817 |  | 3.831E-05 | 0.01468716 |
|---|
| regulation of caspase activity | GO:0043281 |  | 6.22E-05 | 0.02043693 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 6.811E-05 | 0.01958173 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 8.447E-05 | 0.0215863 |
|---|
| response to unfolded protein | GO:0006986 |  | 9.926E-05 | 0.02282952 |
|---|
| response to protein stimulus | GO:0051789 |  | 1.2E-04 | 0.02509127 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.329E-11 | 3.247E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 4.287E-08 | 5.237E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 7.329E-08 | 5.968E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.076E-07 | 1.268E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.118E-07 | 1.035E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 2.487E-07 | 1.013E-04 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 3.207E-07 | 1.119E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 4.054E-07 | 1.238E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.691E-07 | 1.273E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 6.16E-07 | 1.505E-04 |
|---|
| nucleus organization | GO:0006997 |  | 2.985E-06 | 6.629E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.578E-11 | 6.202E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 7.008E-08 | 8.43E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.224E-07 | 9.812E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.409E-07 | 2.051E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.656E-07 | 1.759E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.084E-07 | 1.638E-04 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 5.318E-07 | 1.828E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 6.654E-07 | 2.001E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 7.7E-07 | 2.058E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.011E-06 | 2.432E-04 |
|---|
| nucleus organization | GO:0006997 |  | 4.181E-06 | 9.145E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.329E-11 | 3.247E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 4.287E-08 | 5.237E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 7.329E-08 | 5.968E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.076E-07 | 1.268E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.118E-07 | 1.035E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 2.487E-07 | 1.013E-04 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 3.207E-07 | 1.119E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 4.054E-07 | 1.238E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.691E-07 | 1.273E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 6.16E-07 | 1.505E-04 |
|---|
| nucleus organization | GO:0006997 |  | 2.985E-06 | 6.629E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.578E-11 | 6.202E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 7.008E-08 | 8.43E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.224E-07 | 9.812E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.409E-07 | 2.051E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.656E-07 | 1.759E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.084E-07 | 1.638E-04 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 5.318E-07 | 1.828E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 6.654E-07 | 2.001E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 7.7E-07 | 2.058E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.011E-06 | 2.432E-04 |
|---|
| nucleus organization | GO:0006997 |  | 4.181E-06 | 9.145E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.981E-08 | 1.064E-04 |
|---|
| caspase regulator activity | GO:0043028 |  | 2.754E-06 | 4.918E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.262E-05 | 0.01502496 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in cell cycle arrest | GO:0006977 |  | 1.448E-05 | 0.01292428 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.831E-05 | 0.0130765 |
|---|
| proteasome regulatory particle | GO:0005838 |  | 2.026E-05 | 0.01205556 |
|---|
| sodium:phosphate symporter activity | GO:0005436 |  | 2.026E-05 | 0.01033334 |
|---|
| phosphate transport | GO:0006817 |  | 2.699E-05 | 0.01204847 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 2.699E-05 | 0.01070975 |
|---|
| caspase inhibitor activity | GO:0043027 |  | 2.699E-05 | 9.639E-03 |
|---|
| mitochondrial respiratory chain | GO:0005746 |  | 3.268E-05 | 0.01060879 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.606E-08 | 9.508E-05 |
|---|
| caspase regulator activity | GO:0043028 |  | 1.492E-06 | 2.721E-03 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in cell cycle arrest | GO:0006977 |  | 8.717E-06 | 0.01060044 |
|---|
| activation of caspase activity | GO:0006919 |  | 8.993E-06 | 8.202E-03 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.257E-05 | 9.168E-03 |
|---|
| cerebellar Purkinje cell layer development | GO:0021680 |  | 1.829E-05 | 0.01111778 |
|---|
| sodium:phosphate symporter activity | GO:0005436 |  | 1.829E-05 | 9.53E-03 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 1.829E-05 | 8.338E-03 |
|---|
| cerebellar cortex formation | GO:0021697 |  | 2.437E-05 | 9.877E-03 |
|---|
| phosphate transport | GO:0006817 |  | 2.437E-05 | 8.889E-03 |
|---|
| proteasome regulatory particle | GO:0005838 |  | 2.437E-05 | 8.081E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.578E-11 | 6.202E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 7.008E-08 | 8.43E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.224E-07 | 9.812E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.409E-07 | 2.051E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.656E-07 | 1.759E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.084E-07 | 1.638E-04 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 5.318E-07 | 1.828E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 6.654E-07 | 2.001E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 7.7E-07 | 2.058E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.011E-06 | 2.432E-04 |
|---|
| nucleus organization | GO:0006997 |  | 4.181E-06 | 9.145E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.981E-08 | 6.856E-05 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in cell cycle arrest | GO:0006977 |  | 1.371E-05 | 0.0157705 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.787E-05 | 0.01370377 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.495E-05 | 0.01434893 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 2.876E-05 | 0.01322803 |
|---|
| phosphate transport | GO:0006817 |  | 3.831E-05 | 0.01468716 |
|---|
| regulation of caspase activity | GO:0043281 |  | 6.22E-05 | 0.02043693 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 6.811E-05 | 0.01958173 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 8.447E-05 | 0.0215863 |
|---|
| response to unfolded protein | GO:0006986 |  | 9.926E-05 | 0.02282952 |
|---|
| response to protein stimulus | GO:0051789 |  | 1.2E-04 | 0.02509127 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.981E-08 | 6.856E-05 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in cell cycle arrest | GO:0006977 |  | 1.371E-05 | 0.0157705 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.787E-05 | 0.01370377 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.495E-05 | 0.01434893 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 2.876E-05 | 0.01322803 |
|---|
| phosphate transport | GO:0006817 |  | 3.831E-05 | 0.01468716 |
|---|
| regulation of caspase activity | GO:0043281 |  | 6.22E-05 | 0.02043693 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 6.811E-05 | 0.01958173 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 8.447E-05 | 0.0215863 |
|---|
| response to unfolded protein | GO:0006986 |  | 9.926E-05 | 0.02282952 |
|---|
| response to protein stimulus | GO:0051789 |  | 1.2E-04 | 0.02509127 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.981E-08 | 6.856E-05 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in cell cycle arrest | GO:0006977 |  | 1.371E-05 | 0.0157705 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.787E-05 | 0.01370377 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.495E-05 | 0.01434893 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 2.876E-05 | 0.01322803 |
|---|
| phosphate transport | GO:0006817 |  | 3.831E-05 | 0.01468716 |
|---|
| regulation of caspase activity | GO:0043281 |  | 6.22E-05 | 0.02043693 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 6.811E-05 | 0.01958173 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 8.447E-05 | 0.0215863 |
|---|
| response to unfolded protein | GO:0006986 |  | 9.926E-05 | 0.02282952 |
|---|
| response to protein stimulus | GO:0051789 |  | 1.2E-04 | 0.02509127 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.981E-08 | 6.856E-05 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in cell cycle arrest | GO:0006977 |  | 1.371E-05 | 0.0157705 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.787E-05 | 0.01370377 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.495E-05 | 0.01434893 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 2.876E-05 | 0.01322803 |
|---|
| phosphate transport | GO:0006817 |  | 3.831E-05 | 0.01468716 |
|---|
| regulation of caspase activity | GO:0043281 |  | 6.22E-05 | 0.02043693 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 6.811E-05 | 0.01958173 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 8.447E-05 | 0.0215863 |
|---|
| response to unfolded protein | GO:0006986 |  | 9.926E-05 | 0.02282952 |
|---|
| response to protein stimulus | GO:0051789 |  | 1.2E-04 | 0.02509127 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.736E-08 | 4.119E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.284E-05 | 0.01523739 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in cell cycle arrest | GO:0006977 |  | 1.438E-05 | 0.01137215 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.756E-05 | 0.01041841 |
|---|
| G1 DNA damage checkpoint | GO:0031571 |  | 2.68E-05 | 0.01272144 |
|---|
| regulation of caspase activity | GO:0043281 |  | 4.18E-05 | 0.01653358 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.56E-05 | 0.01545936 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 5.607E-05 | 0.01663285 |
|---|
| phosphate transport | GO:0006817 |  | 6.303E-05 | 0.01661886 |
|---|
| G1/S transition checkpoint | GO:0031575 |  | 1.001E-04 | 0.02375216 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.143E-04 | 0.02466268 |
|---|



