Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 10907-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2723 | 1.917e-02 | 2.298e-02 | 1.595e-01 |
|---|
| IPC-NIBC-129 | 0.2575 | 3.761e-02 | 4.698e-02 | 3.713e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2166 | 3.727e-03 | 6.336e-03 | 1.401e-01 |
|---|
| Loi_GPL570 | 0.2890 | 5.546e-02 | 5.506e-02 | 2.749e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1508 | 9.700e-05 | 4.840e-04 | 2.068e-03 |
|---|
| Parker_GPL1390 | 0.2789 | 3.529e-02 | 3.181e-02 | 3.088e-01 |
|---|
| Parker_GPL887 | 0.1707 | 4.278e-01 | 4.625e-01 | 4.521e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3748 | 8.545e-01 | 8.515e-01 | 9.997e-01 |
|---|
| Schmidt | 0.2061 | 5.264e-03 | 7.727e-03 | 7.842e-02 |
|---|
| Sotiriou | 0.3542 | 3.242e-03 | 1.004e-03 | 7.510e-02 |
|---|
| Wang | 0.2004 | 1.284e-01 | 9.207e-02 | 4.288e-01 |
|---|
| Zhang | 0.0356 | 7.805e-02 | 4.868e-02 | 1.924e-01 |
|---|
| Zhou | 0.3972 | 5.322e-02 | 5.552e-02 | 3.353e-01 |
|---|
Expression data for subnetwork 10907-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
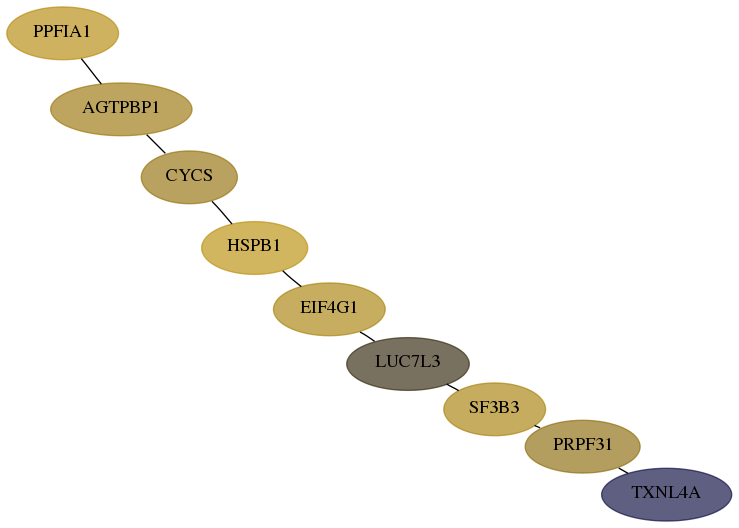
Score for each gene in subnetwork 10907-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| agtpbp1 |   | 11 | 21 | 17 | 13 | 0.086 | 0.219 | 0.009 | 0.181 | 0.071 | 0.078 | 0.296 | 0.026 | 0.110 | 0.151 | 0.103 | 0.064 | 0.226 |
|---|
| prpf31 |   | 1 | 164 | 251 | 254 | 0.060 | 0.048 | 0.113 | 0.024 | 0.121 | 0.070 | 0.194 | 0.146 | 0.111 | 0.135 | 0.083 | -0.048 | 0.295 |
|---|
| cycs |   | 10 | 25 | 17 | 14 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | -0.056 | 0.202 | 0.272 | 0.157 | 0.077 | 0.190 | 0.135 | 0.234 |
|---|
| luc7l3 |   | 7 | 38 | 1 | 5 | 0.006 | 0.111 | -0.046 | -0.016 | 0.121 | 0.071 | -0.318 | -0.084 | 0.159 | 0.098 | 0.155 | 0.067 | 0.065 |
|---|
| ppfia1 |   | 8 | 33 | 17 | 16 | 0.164 | 0.080 | 0.149 | 0.269 | 0.151 | 0.214 | 0.093 | 0.225 | 0.036 | 0.176 | 0.153 | 0.127 | 0.272 |
|---|
| eif4g1 |   | 5 | 50 | 153 | 142 | 0.123 | 0.262 | 0.043 | 0.013 | 0.098 | 0.189 | 0.016 | 0.194 | 0.076 | 0.147 | -0.154 | -0.028 | 0.268 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| sf3b3 |   | 7 | 38 | 1 | 5 | 0.119 | 0.241 | 0.143 | -0.019 | 0.082 | 0.222 | 0.007 | 0.271 | 0.168 | 0.060 | 0.017 | -0.077 | 0.372 |
|---|
| txnl4a |   | 2 | 104 | 251 | 239 | -0.008 | 0.172 | 0.148 | 0.211 | 0.199 | 0.132 | 0.270 | 0.207 | 0.056 | 0.194 | 0.015 | 0.082 | 0.135 |
|---|
GO Enrichment output for subnetwork 10907-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.182E-05 | 0.02719098 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.266E-05 | 0.01455722 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.498E-05 | 0.01148687 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.043E-04 | 0.05997574 |
|---|
| regulation of translation | GO:0006417 |  | 1.392E-04 | 0.06404491 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.695E-04 | 0.0649578 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.695E-04 | 0.05567811 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.078E-04 | 0.0597566 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.285E-04 | 0.05839304 |
|---|
| spliceosome assembly | GO:0000245 |  | 4.282E-04 | 0.09848312 |
|---|
| response to heat | GO:0009408 |  | 4.282E-04 | 0.08953011 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.15E-08 | 2.81E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.391E-07 | 1.7E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.717E-07 | 2.213E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.145E-07 | 1.921E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.13E-07 | 2.018E-04 |
|---|
| nucleus organization | GO:0006997 |  | 2.003E-06 | 8.156E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 3.496E-06 | 1.22E-03 |
|---|
| assembly of spliceosomal tri-snRNP | GO:0000244 |  | 3.653E-06 | 1.115E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.958E-06 | 1.074E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 4.459E-06 | 1.089E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 5.001E-06 | 1.111E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.173E-08 | 5.229E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.627E-07 | 3.16E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 5.128E-07 | 4.113E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 5.935E-07 | 3.57E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 7.792E-07 | 3.749E-04 |
|---|
| nucleus organization | GO:0006997 |  | 3.225E-06 | 1.293E-03 |
|---|
| cellular component disassembly | GO:0022411 |  | 6.579E-06 | 2.261E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 6.579E-06 | 1.979E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 7.005E-06 | 1.873E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 8.889E-06 | 2.139E-03 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 9.945E-06 | 2.175E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.15E-08 | 2.81E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.391E-07 | 1.7E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.717E-07 | 2.213E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 3.145E-07 | 1.921E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 4.13E-07 | 2.018E-04 |
|---|
| nucleus organization | GO:0006997 |  | 2.003E-06 | 8.156E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 3.496E-06 | 1.22E-03 |
|---|
| assembly of spliceosomal tri-snRNP | GO:0000244 |  | 3.653E-06 | 1.115E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.958E-06 | 1.074E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 4.459E-06 | 1.089E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 5.001E-06 | 1.111E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.173E-08 | 5.229E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.627E-07 | 3.16E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 5.128E-07 | 4.113E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 5.935E-07 | 3.57E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 7.792E-07 | 3.749E-04 |
|---|
| nucleus organization | GO:0006997 |  | 3.225E-06 | 1.293E-03 |
|---|
| cellular component disassembly | GO:0022411 |  | 6.579E-06 | 2.261E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 6.579E-06 | 1.979E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 7.005E-06 | 1.873E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 8.889E-06 | 2.139E-03 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 9.945E-06 | 2.175E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| RNA splicing factor activity. transesterification mechanism | GO:0031202 |  | 3.169E-07 | 1.132E-03 |
|---|
| small nuclear ribonucleoprotein complex | GO:0030532 |  | 3.601E-07 | 6.429E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.661E-06 | 1.977E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 6.954E-06 | 6.208E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 7.743E-06 | 5.53E-03 |
|---|
| Cajal body | GO:0015030 |  | 9.269E-06 | 5.517E-03 |
|---|
| nuclear speck | GO:0016607 |  | 1.127E-05 | 5.75E-03 |
|---|
| eukaryotic translation initiation factor 4F complex | GO:0016281 |  | 3.007E-05 | 0.0134218 |
|---|
| ribonucleoprotein binding | GO:0043021 |  | 3.468E-05 | 0.01376158 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 4.489E-05 | 0.01603191 |
|---|
| protein phosphatase type 2A complex | GO:0000159 |  | 5.049E-05 | 0.01639108 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| RNA splicing factor activity. transesterification mechanism | GO:0031202 |  | 3.034E-07 | 1.107E-03 |
|---|
| small nuclear ribonucleoprotein complex | GO:0030532 |  | 3.034E-07 | 5.535E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.259E-06 | 1.531E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.711E-06 | 1.561E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 6.582E-06 | 4.802E-03 |
|---|
| cerebellar Purkinje cell layer development | GO:0021680 |  | 6.582E-06 | 4.002E-03 |
|---|
| Cajal body | GO:0015030 |  | 6.582E-06 | 3.43E-03 |
|---|
| cerebellar cortex formation | GO:0021697 |  | 8.773E-06 | 4E-03 |
|---|
| eukaryotic translation initiation factor 4F complex | GO:0016281 |  | 8.773E-06 | 3.556E-03 |
|---|
| cell differentiation in hindbrain | GO:0021533 |  | 1.128E-05 | 4.113E-03 |
|---|
| nuclear speck | GO:0016607 |  | 1.352E-05 | 4.483E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.173E-08 | 5.229E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.627E-07 | 3.16E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 5.128E-07 | 4.113E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 5.935E-07 | 3.57E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 7.792E-07 | 3.749E-04 |
|---|
| nucleus organization | GO:0006997 |  | 3.225E-06 | 1.293E-03 |
|---|
| cellular component disassembly | GO:0022411 |  | 6.579E-06 | 2.261E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 6.579E-06 | 1.979E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 7.005E-06 | 1.873E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 8.889E-06 | 2.139E-03 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 9.945E-06 | 2.175E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.182E-05 | 0.02719098 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.266E-05 | 0.01455722 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.498E-05 | 0.01148687 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.043E-04 | 0.05997574 |
|---|
| regulation of translation | GO:0006417 |  | 1.392E-04 | 0.06404491 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.695E-04 | 0.0649578 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.695E-04 | 0.05567811 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.078E-04 | 0.0597566 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.285E-04 | 0.05839304 |
|---|
| spliceosome assembly | GO:0000245 |  | 4.282E-04 | 0.09848312 |
|---|
| response to heat | GO:0009408 |  | 4.282E-04 | 0.08953011 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.182E-05 | 0.02719098 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.266E-05 | 0.01455722 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.498E-05 | 0.01148687 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.043E-04 | 0.05997574 |
|---|
| regulation of translation | GO:0006417 |  | 1.392E-04 | 0.06404491 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.695E-04 | 0.0649578 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.695E-04 | 0.05567811 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.078E-04 | 0.0597566 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.285E-04 | 0.05839304 |
|---|
| spliceosome assembly | GO:0000245 |  | 4.282E-04 | 0.09848312 |
|---|
| response to heat | GO:0009408 |  | 4.282E-04 | 0.08953011 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.182E-05 | 0.02719098 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.266E-05 | 0.01455722 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.498E-05 | 0.01148687 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.043E-04 | 0.05997574 |
|---|
| regulation of translation | GO:0006417 |  | 1.392E-04 | 0.06404491 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.695E-04 | 0.0649578 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.695E-04 | 0.05567811 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.078E-04 | 0.0597566 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.285E-04 | 0.05839304 |
|---|
| spliceosome assembly | GO:0000245 |  | 4.282E-04 | 0.09848312 |
|---|
| response to heat | GO:0009408 |  | 4.282E-04 | 0.08953011 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.182E-05 | 0.02719098 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.266E-05 | 0.01455722 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.498E-05 | 0.01148687 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.043E-04 | 0.05997574 |
|---|
| regulation of translation | GO:0006417 |  | 1.392E-04 | 0.06404491 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.695E-04 | 0.0649578 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.695E-04 | 0.05567811 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.078E-04 | 0.0597566 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.285E-04 | 0.05839304 |
|---|
| spliceosome assembly | GO:0000245 |  | 4.282E-04 | 0.09848312 |
|---|
| response to heat | GO:0009408 |  | 4.282E-04 | 0.08953011 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translational initiation | GO:0006446 |  | 9.222E-06 | 0.02188438 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.158E-05 | 0.01374004 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.5E-05 | 0.01186579 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 9.215E-05 | 0.05466615 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.456E-04 | 0.06909541 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.608E-04 | 0.06360639 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.768E-04 | 0.05993959 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.679E-04 | 0.07947904 |
|---|
| response to heat | GO:0009408 |  | 3.315E-04 | 0.08741455 |
|---|
| spliceosome assembly | GO:0000245 |  | 4.786E-04 | 0.11357447 |
|---|
| nucleus organization | GO:0006997 |  | 5.057E-04 | 0.109091 |
|---|



