Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 10799-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1889 | 8.778e-03 | 1.048e-02 | 8.983e-02 |
|---|
| IPC-NIBC-129 | 0.2480 | 4.659e-02 | 5.904e-02 | 4.315e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2546 | 1.420e-02 | 2.110e-02 | 3.228e-01 |
|---|
| Loi_GPL570 | 0.2265 | 6.316e-02 | 6.273e-02 | 3.020e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1607 | 1.000e-05 | 6.800e-05 | 8.300e-05 |
|---|
| Parker_GPL1390 | 0.2634 | 1.146e-01 | 1.155e-01 | 5.767e-01 |
|---|
| Parker_GPL887 | 0.0279 | 9.241e-01 | 9.325e-01 | 9.585e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3624 | 8.451e-01 | 8.413e-01 | 9.996e-01 |
|---|
| Schmidt | 0.2384 | 2.017e-02 | 2.791e-02 | 2.202e-01 |
|---|
| Sotiriou | 0.2722 | 8.742e-03 | 3.503e-03 | 1.486e-01 |
|---|
| Wang | 0.1639 | 1.095e-01 | 7.647e-02 | 3.876e-01 |
|---|
| Zhang | 0.0373 | 1.318e-01 | 9.113e-02 | 3.055e-01 |
|---|
| Zhou | 0.3370 | 5.994e-02 | 6.326e-02 | 3.718e-01 |
|---|
Expression data for subnetwork 10799-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
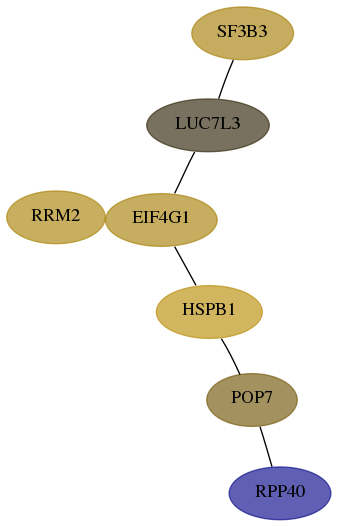
Score for each gene in subnetwork 10799-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| pop7 |   | 1 | 164 | 405 | 405 | 0.032 | 0.154 | 0.110 | 0.147 | 0.123 | 0.019 | 0.443 | 0.217 | 0.097 | 0.090 | 0.127 | 0.107 | 0.080 |
|---|
| rpp40 |   | 1 | 164 | 405 | 405 | -0.059 | 0.190 | 0.138 | 0.150 | 0.104 | 0.207 | -0.229 | 0.156 | 0.219 | 0.077 | -0.026 | 0.014 | -0.126 |
|---|
| sf3b3 |   | 7 | 38 | 1 | 5 | 0.119 | 0.241 | 0.143 | -0.019 | 0.082 | 0.222 | 0.007 | 0.271 | 0.168 | 0.060 | 0.017 | -0.077 | 0.372 |
|---|
| rrm2 |   | 21 | 10 | 44 | 26 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | 0.117 | 0.129 | 0.302 |
|---|
| luc7l3 |   | 7 | 38 | 1 | 5 | 0.006 | 0.111 | -0.046 | -0.016 | 0.121 | 0.071 | -0.318 | -0.084 | 0.159 | 0.098 | 0.155 | 0.067 | 0.065 |
|---|
| eif4g1 |   | 5 | 50 | 153 | 142 | 0.123 | 0.262 | 0.043 | 0.013 | 0.098 | 0.189 | 0.016 | 0.194 | 0.076 | 0.147 | -0.154 | -0.028 | 0.268 |
|---|
GO Enrichment output for subnetwork 10799-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.703E-09 | 8.516E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.478E-09 | 7.449E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.45E-07 | 1.878E-04 |
|---|
| tRNA processing | GO:0008033 |  | 2.256E-06 | 1.297E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.592E-06 | 1.192E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 7.924E-06 | 3.038E-03 |
|---|
| tRNA metabolic process | GO:0006399 |  | 1.271E-05 | 4.175E-03 |
|---|
| regulation of translation | GO:0006417 |  | 2.898E-05 | 8.333E-03 |
|---|
| response to heat | GO:0009408 |  | 1.551E-04 | 0.03963101 |
|---|
| response to temperature stimulus | GO:0009266 |  | 3.997E-04 | 0.0919208 |
|---|
| response to unfolded protein | GO:0006986 |  | 1E-03 | 0.20911357 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translational initiation | GO:0006446 |  | 7.221E-07 | 1.764E-03 |
|---|
| tRNA processing | GO:0008033 |  | 1.099E-06 | 1.343E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.192E-06 | 1.785E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 3.757E-06 | 2.294E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.633E-05 | 0.01286521 |
|---|
| response to heat | GO:0009408 |  | 4.832E-05 | 0.01967507 |
|---|
| response to temperature stimulus | GO:0009266 |  | 1.32E-04 | 0.04606779 |
|---|
| protein oligomerization | GO:0051259 |  | 3.173E-04 | 0.09689608 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.116E-04 | 0.11172967 |
|---|
| response to protein stimulus | GO:0051789 |  | 4.681E-04 | 0.11436276 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.687E-03 | 0.37466827 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.197E-09 | 5.285E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 5.269E-09 | 6.339E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 9.629E-08 | 7.723E-05 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.453E-06 | 8.743E-04 |
|---|
| tRNA processing | GO:0008033 |  | 2.229E-06 | 1.072E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 4.691E-06 | 1.881E-03 |
|---|
| response to heat | GO:0009408 |  | 7.431E-05 | 0.02554119 |
|---|
| response to temperature stimulus | GO:0009266 |  | 2E-04 | 0.06013926 |
|---|
| response to unfolded protein | GO:0006986 |  | 6.017E-04 | 0.16086449 |
|---|
| response to protein stimulus | GO:0051789 |  | 6.892E-04 | 0.16582656 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.157E-03 | 0.47169196 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translational initiation | GO:0006446 |  | 7.221E-07 | 1.764E-03 |
|---|
| tRNA processing | GO:0008033 |  | 1.099E-06 | 1.343E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.192E-06 | 1.785E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 3.757E-06 | 2.294E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.633E-05 | 0.01286521 |
|---|
| response to heat | GO:0009408 |  | 4.832E-05 | 0.01967507 |
|---|
| response to temperature stimulus | GO:0009266 |  | 1.32E-04 | 0.04606779 |
|---|
| protein oligomerization | GO:0051259 |  | 3.173E-04 | 0.09689608 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.116E-04 | 0.11172967 |
|---|
| response to protein stimulus | GO:0051789 |  | 4.681E-04 | 0.11436276 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.687E-03 | 0.37466827 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.197E-09 | 5.285E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 5.269E-09 | 6.339E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 9.629E-08 | 7.723E-05 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.453E-06 | 8.743E-04 |
|---|
| tRNA processing | GO:0008033 |  | 2.229E-06 | 1.072E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 4.691E-06 | 1.881E-03 |
|---|
| response to heat | GO:0009408 |  | 7.431E-05 | 0.02554119 |
|---|
| response to temperature stimulus | GO:0009266 |  | 2E-04 | 0.06013926 |
|---|
| response to unfolded protein | GO:0006986 |  | 6.017E-04 | 0.16086449 |
|---|
| response to protein stimulus | GO:0051789 |  | 6.892E-04 | 0.16582656 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.157E-03 | 0.47169196 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleolar ribonuclease P complex | GO:0005655 |  | 1.313E-09 | 4.687E-06 |
|---|
| ribonuclease P activity | GO:0004526 |  | 7.869E-09 | 1.405E-05 |
|---|
| tRNA-specific ribonuclease activity | GO:0004549 |  | 1.082E-08 | 1.288E-05 |
|---|
| nucleolar part | GO:0044452 |  | 1.874E-08 | 1.673E-05 |
|---|
| endoribonuclease activity. producing 5'-phosphomonoesters | GO:0016891 |  | 1.007E-07 | 7.195E-05 |
|---|
| endonuclease activity. active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters | GO:0016893 |  | 1.699E-07 | 1.011E-04 |
|---|
| endoribonuclease activity | GO:0004521 |  | 3.561E-07 | 1.817E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 6.946E-07 | 3.101E-04 |
|---|
| ribonuclease activity | GO:0004540 |  | 1.199E-06 | 4.756E-04 |
|---|
| tRNA processing | GO:0008033 |  | 1.612E-06 | 5.756E-04 |
|---|
| oxidoreductase activity. acting on CH or CH2 groups | GO:0016725 |  | 4.058E-06 | 1.318E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleolar ribonuclease P complex | GO:0005655 |  | 1.024E-09 | 3.735E-06 |
|---|
| ribonuclease P activity | GO:0004526 |  | 6.139E-09 | 1.12E-05 |
|---|
| tRNA-specific ribonuclease activity | GO:0004549 |  | 1.125E-08 | 1.368E-05 |
|---|
| nucleolar part | GO:0044452 |  | 1.462E-08 | 1.333E-05 |
|---|
| endoribonuclease activity. producing 5'-phosphomonoesters | GO:0016891 |  | 7.859E-08 | 5.734E-05 |
|---|
| endonuclease activity. active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters | GO:0016893 |  | 1.326E-07 | 8.061E-05 |
|---|
| endoribonuclease activity | GO:0004521 |  | 3.953E-07 | 2.06E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 4.291E-07 | 1.957E-04 |
|---|
| ribonuclease activity | GO:0004540 |  | 1.405E-06 | 5.695E-04 |
|---|
| tRNA processing | GO:0008033 |  | 2.108E-06 | 7.691E-04 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.395E-06 | 7.943E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.197E-09 | 5.285E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 5.269E-09 | 6.339E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 9.629E-08 | 7.723E-05 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.453E-06 | 8.743E-04 |
|---|
| tRNA processing | GO:0008033 |  | 2.229E-06 | 1.072E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 4.691E-06 | 1.881E-03 |
|---|
| response to heat | GO:0009408 |  | 7.431E-05 | 0.02554119 |
|---|
| response to temperature stimulus | GO:0009266 |  | 2E-04 | 0.06013926 |
|---|
| response to unfolded protein | GO:0006986 |  | 6.017E-04 | 0.16086449 |
|---|
| response to protein stimulus | GO:0051789 |  | 6.892E-04 | 0.16582656 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.157E-03 | 0.47169196 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.703E-09 | 8.516E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.478E-09 | 7.449E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.45E-07 | 1.878E-04 |
|---|
| tRNA processing | GO:0008033 |  | 2.256E-06 | 1.297E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.592E-06 | 1.192E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 7.924E-06 | 3.038E-03 |
|---|
| tRNA metabolic process | GO:0006399 |  | 1.271E-05 | 4.175E-03 |
|---|
| regulation of translation | GO:0006417 |  | 2.898E-05 | 8.333E-03 |
|---|
| response to heat | GO:0009408 |  | 1.551E-04 | 0.03963101 |
|---|
| response to temperature stimulus | GO:0009266 |  | 3.997E-04 | 0.0919208 |
|---|
| response to unfolded protein | GO:0006986 |  | 1E-03 | 0.20911357 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.703E-09 | 8.516E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.478E-09 | 7.449E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.45E-07 | 1.878E-04 |
|---|
| tRNA processing | GO:0008033 |  | 2.256E-06 | 1.297E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.592E-06 | 1.192E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 7.924E-06 | 3.038E-03 |
|---|
| tRNA metabolic process | GO:0006399 |  | 1.271E-05 | 4.175E-03 |
|---|
| regulation of translation | GO:0006417 |  | 2.898E-05 | 8.333E-03 |
|---|
| response to heat | GO:0009408 |  | 1.551E-04 | 0.03963101 |
|---|
| response to temperature stimulus | GO:0009266 |  | 3.997E-04 | 0.0919208 |
|---|
| response to unfolded protein | GO:0006986 |  | 1E-03 | 0.20911357 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.703E-09 | 8.516E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.478E-09 | 7.449E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.45E-07 | 1.878E-04 |
|---|
| tRNA processing | GO:0008033 |  | 2.256E-06 | 1.297E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.592E-06 | 1.192E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 7.924E-06 | 3.038E-03 |
|---|
| tRNA metabolic process | GO:0006399 |  | 1.271E-05 | 4.175E-03 |
|---|
| regulation of translation | GO:0006417 |  | 2.898E-05 | 8.333E-03 |
|---|
| response to heat | GO:0009408 |  | 1.551E-04 | 0.03963101 |
|---|
| response to temperature stimulus | GO:0009266 |  | 3.997E-04 | 0.0919208 |
|---|
| response to unfolded protein | GO:0006986 |  | 1E-03 | 0.20911357 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.703E-09 | 8.516E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.478E-09 | 7.449E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.45E-07 | 1.878E-04 |
|---|
| tRNA processing | GO:0008033 |  | 2.256E-06 | 1.297E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.592E-06 | 1.192E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 7.924E-06 | 3.038E-03 |
|---|
| tRNA metabolic process | GO:0006399 |  | 1.271E-05 | 4.175E-03 |
|---|
| regulation of translation | GO:0006417 |  | 2.898E-05 | 8.333E-03 |
|---|
| response to heat | GO:0009408 |  | 1.551E-04 | 0.03963101 |
|---|
| response to temperature stimulus | GO:0009266 |  | 3.997E-04 | 0.0919208 |
|---|
| response to unfolded protein | GO:0006986 |  | 1E-03 | 0.20911357 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.4E-09 | 1.044E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.038E-09 | 8.351E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.927E-07 | 1.525E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.886E-06 | 1.119E-03 |
|---|
| tRNA processing | GO:0008033 |  | 2.148E-06 | 1.019E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 5.651E-06 | 2.235E-03 |
|---|
| tRNA metabolic process | GO:0006399 |  | 1.301E-05 | 4.41E-03 |
|---|
| response to heat | GO:0009408 |  | 1.2E-04 | 0.03558441 |
|---|
| response to temperature stimulus | GO:0009266 |  | 3.225E-04 | 0.08502278 |
|---|
| response to unfolded protein | GO:0006986 |  | 8.584E-04 | 0.20370181 |
|---|
| response to protein stimulus | GO:0051789 |  | 9.683E-04 | 0.20889502 |
|---|



