Study VanDeVijver-ER-pos
Study informations
14 subnetworks in total page | file
272 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 83
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.5273 | 0.000e+00 | 0.000e+00 | 2.660e-04 | 0.000e+00 |
|---|
| IPC | 0.4586 | 1.077e-01 | 8.835e-02 | 9.575e-01 | 9.109e-03 |
|---|
| Loi | 0.5088 | 0.000e+00 | 0.000e+00 | 2.577e-03 | 0.000e+00 |
|---|
| Schmidt | 0.2936 | 1.381e-02 | 3.800e-03 | 2.899e-01 | 1.522e-05 |
|---|
| Wang | 0.3377 | 1.350e-04 | 3.000e-06 | 5.216e-02 | 2.112e-11 |
|---|
Expression data for subnetwork 83 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
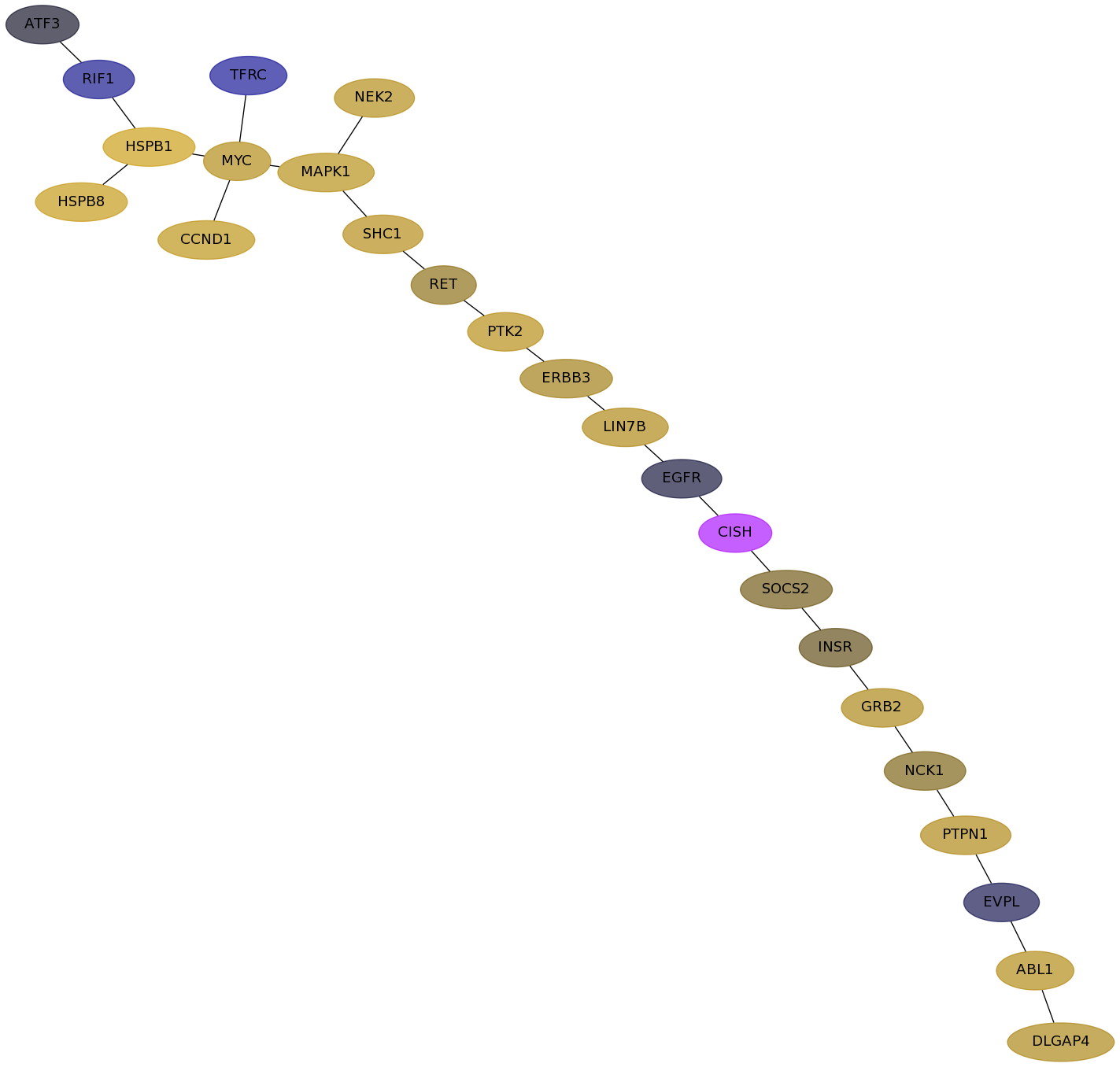
Score for each gene in subnetwork 83 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| NCK1 |   | 3 | 13 | 80 | 84 | 0.037 | 0.127 | 0.044 | 0.103 | 0.055 |
|---|
| RIF1 |   | 3 | 13 | 1 | 5 | -0.055 | 0.053 | -0.056 | -0.038 | 0.085 |
|---|
| RET |   | 4 | 6 | 167 | 167 | 0.055 | -0.063 | 0.119 | 0.041 | 0.006 |
|---|
| SOCS2 |   | 1 | 59 | 179 | 179 | 0.026 | -0.106 | -0.043 | -0.118 | -0.158 |
|---|
| DLGAP4 |   | 1 | 59 | 179 | 179 | 0.122 | -0.078 | 0.149 | 0.014 | 0.021 |
|---|
| ATF3 |   | 4 | 6 | 80 | 82 | -0.003 | -0.016 | 0.092 | -0.045 | -0.079 |
|---|
| PTK2 |   | 1 | 59 | 179 | 179 | 0.153 | 0.071 | 0.134 | 0.108 | 0.173 |
|---|
| EVPL |   | 3 | 13 | 80 | 84 | -0.011 | 0.035 | 0.178 | 0.232 | -0.047 |
|---|
| EGFR |   | 1 | 59 | 179 | 179 | -0.006 | -0.114 | 0.012 | -0.027 | -0.136 |
|---|
| TFRC |   | 1 | 59 | 179 | 179 | -0.068 | 0.015 | 0.148 | 0.159 | 0.087 |
|---|
| GRB2 |   | 7 | 1 | 80 | 80 | 0.121 | 0.037 | 0.087 | 0.182 | -0.037 |
|---|
| CCND1 |   | 4 | 6 | 36 | 36 | 0.189 | 0.059 | 0.145 | 0.009 | 0.146 |
|---|
| ABL1 |   | 1 | 59 | 179 | 179 | 0.142 | -0.042 | -0.056 | -0.127 | -0.078 |
|---|
| CISH |   | 1 | 59 | 179 | 179 | undef | 0.045 | 0.151 | undef | undef |
|---|
| INSR |   | 1 | 59 | 179 | 179 | 0.017 | 0.043 | -0.052 | -0.059 | -0.048 |
|---|
| LIN7B |   | 1 | 59 | 179 | 179 | 0.130 | 0.158 | 0.181 | 0.214 | 0.130 |
|---|
| PTPN1 |   | 1 | 59 | 179 | 179 | 0.129 | 0.043 | 0.140 | 0.055 | -0.033 |
|---|
| SHC1 |   | 4 | 6 | 167 | 167 | 0.149 | -0.137 | 0.092 | -0.200 | 0.113 |
|---|
| MYC |   | 4 | 6 | 1 | 3 | 0.142 | 0.031 | 0.041 | 0.035 | -0.138 |
|---|
| HSPB8 |   | 3 | 13 | 1 | 5 | 0.231 | 0.010 | 0.076 | -0.044 | 0.013 |
|---|
| NEK2 |   | 3 | 13 | 110 | 111 | 0.147 | 0.147 | 0.232 | 0.245 | 0.336 |
|---|
| ERBB3 |   | 1 | 59 | 179 | 179 | 0.090 | -0.014 | 0.060 | -0.110 | -0.025 |
|---|
| MAPK1 |   | 5 | 4 | 110 | 110 | 0.157 | -0.023 | -0.125 | -0.115 | 0.092 |
|---|
| HSPB1 |   | 6 | 2 | 1 | 1 | 0.262 | 0.053 | 0.231 | 0.043 | 0.144 |
|---|
GO Enrichment output for subnetwork 83 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.953E-07 | 6.791E-04 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 4.14E-07 | 4.761E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.483E-07 | 3.437E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 8.92E-07 | 5.129E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.266E-06 | 5.825E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 3.318E-06 | 1.272E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.991E-06 | 1.311E-03 |
|---|
| signal complex assembly | GO:0007172 |  | 8.565E-06 | 2.462E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 8.565E-06 | 2.189E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.036E-05 | 2.384E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.429E-05 | 2.987E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.656E-07 | 1.137E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 5.422E-07 | 6.623E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.818E-07 | 5.552E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.741E-06 | 1.063E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.92E-06 | 9.382E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 4.566E-06 | 1.859E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.973E-06 | 1.736E-03 |
|---|
| stem cell development | GO:0048864 |  | 6.726E-06 | 2.054E-03 |
|---|
| signal complex assembly | GO:0007172 |  | 8.154E-06 | 2.213E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 9.768E-06 | 2.386E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.578E-05 | 3.505E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.313E-07 | 1.038E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 5.054E-07 | 6.08E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.274E-07 | 5.833E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.603E-06 | 9.645E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.868E-06 | 8.988E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 4.541E-06 | 1.821E-03 |
|---|
| stem cell development | GO:0048864 |  | 4.679E-06 | 1.608E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.395E-06 | 1.622E-03 |
|---|
| signal complex assembly | GO:0007172 |  | 8.697E-06 | 2.325E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 8.697E-06 | 2.092E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.413E-05 | 3.091E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.953E-07 | 6.791E-04 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 4.14E-07 | 4.761E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.483E-07 | 3.437E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 8.92E-07 | 5.129E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.266E-06 | 5.825E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 3.318E-06 | 1.272E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.991E-06 | 1.311E-03 |
|---|
| signal complex assembly | GO:0007172 |  | 8.565E-06 | 2.462E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 8.565E-06 | 2.189E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.036E-05 | 2.384E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.429E-05 | 2.987E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.953E-07 | 6.791E-04 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 4.14E-07 | 4.761E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.483E-07 | 3.437E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 8.92E-07 | 5.129E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.266E-06 | 5.825E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 3.318E-06 | 1.272E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.991E-06 | 1.311E-03 |
|---|
| signal complex assembly | GO:0007172 |  | 8.565E-06 | 2.462E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 8.565E-06 | 2.189E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.036E-05 | 2.384E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.429E-05 | 2.987E-03 |
|---|


