Study VanDeVijver-ER-pos
Study informations
14 subnetworks in total page | file
272 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 82
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.5270 | 0.000e+00 | 0.000e+00 | 2.700e-04 | 0.000e+00 |
|---|
| IPC | 0.4644 | 1.012e-01 | 8.183e-02 | 9.569e-01 | 7.924e-03 |
|---|
| Loi | 0.5078 | 0.000e+00 | 0.000e+00 | 2.638e-03 | 0.000e+00 |
|---|
| Schmidt | 0.2956 | 1.290e-02 | 3.463e-03 | 2.785e-01 | 1.244e-05 |
|---|
| Wang | 0.3402 | 1.160e-04 | 3.000e-06 | 4.728e-02 | 1.645e-11 |
|---|
Expression data for subnetwork 82 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
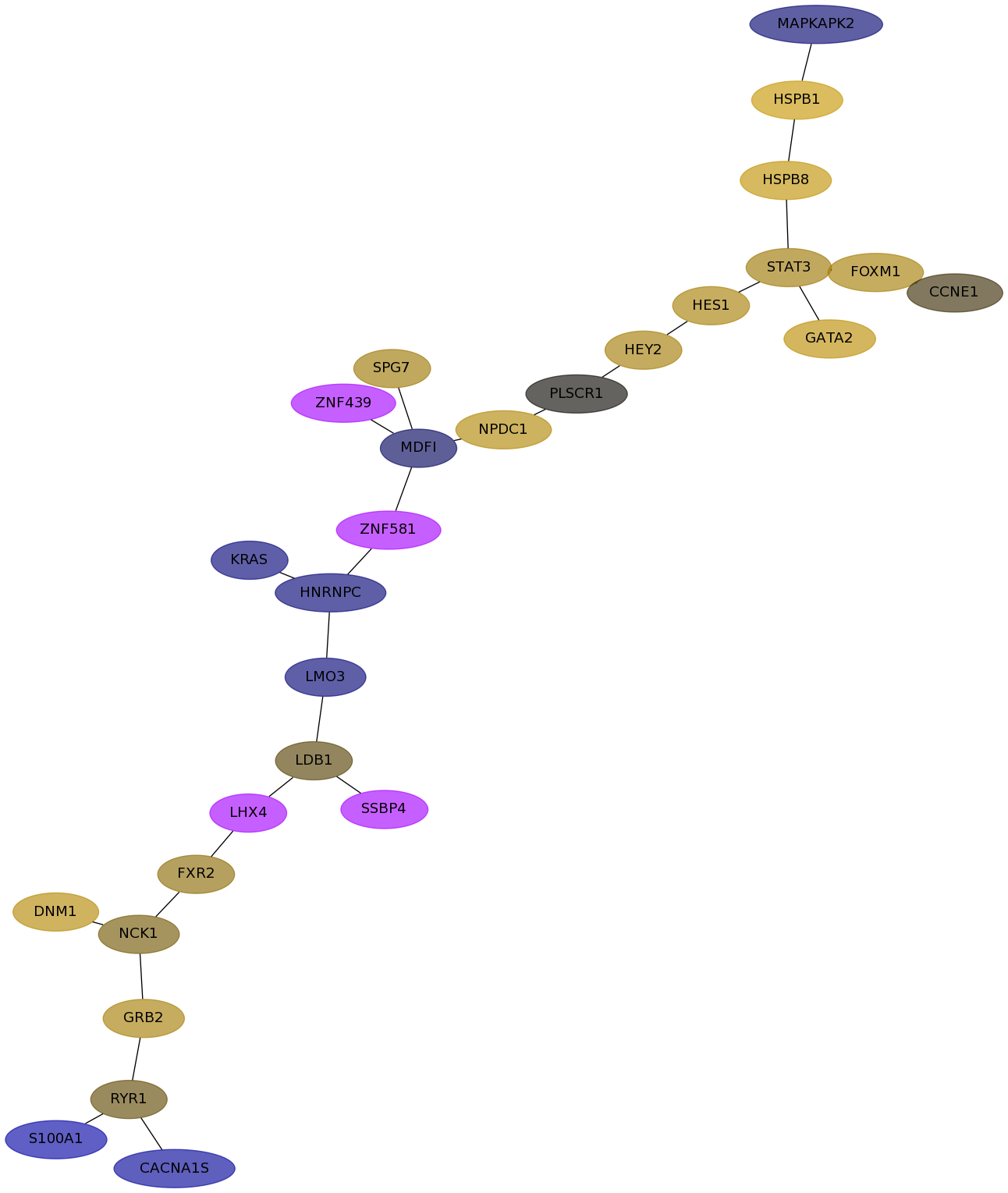
Score for each gene in subnetwork 82 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| HES1 |   | 1 | 59 | 148 | 150 | 0.124 | -0.037 | -0.022 | 0.031 | -0.023 |
|---|
| DNM1 |   | 1 | 59 | 148 | 150 | 0.168 | -0.092 | -0.126 | -0.082 | 0.161 |
|---|
| LMO3 |   | 1 | 59 | 148 | 150 | -0.038 | -0.076 | 0.075 | 0.059 | -0.035 |
|---|
| ZNF439 |   | 1 | 59 | 148 | 150 | undef | -0.014 | 0.077 | undef | undef |
|---|
| PLSCR1 |   | 1 | 59 | 148 | 150 | 0.001 | -0.065 | 0.104 | 0.124 | -0.134 |
|---|
| RYR1 |   | 1 | 59 | 148 | 150 | 0.023 | -0.111 | -0.018 | 0.162 | 0.072 |
|---|
| NCK1 |   | 3 | 13 | 80 | 84 | 0.037 | 0.127 | 0.044 | 0.103 | 0.055 |
|---|
| FOXM1 |   | 2 | 27 | 110 | 113 | 0.119 | 0.137 | 0.170 | 0.219 | 0.183 |
|---|
| LDB1 |   | 1 | 59 | 148 | 150 | 0.017 | 0.000 | -0.015 | 0.214 | -0.137 |
|---|
| HNRNPC |   | 1 | 59 | 148 | 150 | -0.038 | -0.131 | 0.097 | 0.133 | 0.164 |
|---|
| KRAS |   | 2 | 27 | 1 | 10 | -0.041 | 0.147 | 0.088 | 0.259 | 0.018 |
|---|
| FXR2 |   | 1 | 59 | 148 | 150 | 0.066 | -0.174 | -0.032 | 0.088 | -0.151 |
|---|
| NPDC1 |   | 1 | 59 | 148 | 150 | 0.160 | -0.160 | 0.031 | -0.080 | 0.029 |
|---|
| CCNE1 |   | 1 | 59 | 148 | 150 | 0.008 | 0.179 | 0.119 | 0.305 | 0.105 |
|---|
| GRB2 |   | 7 | 1 | 80 | 80 | 0.121 | 0.037 | 0.087 | 0.182 | -0.037 |
|---|
| SPG7 |   | 1 | 59 | 148 | 150 | 0.100 | -0.077 | 0.157 | -0.069 | -0.007 |
|---|
| MAPKAPK2 |   | 1 | 59 | 148 | 150 | -0.033 | -0.049 | 0.100 | 0.210 | 0.101 |
|---|
| HEY2 |   | 1 | 59 | 148 | 150 | 0.113 | -0.042 | 0.164 | -0.002 | 0.119 |
|---|
| ZNF581 |   | 2 | 27 | 148 | 148 | undef | -0.087 | 0.146 | undef | undef |
|---|
| LHX4 |   | 2 | 27 | 80 | 87 | undef | 0.205 | 0.125 | undef | undef |
|---|
| GATA2 |   | 4 | 6 | 80 | 82 | 0.201 | -0.014 | 0.121 | 0.080 | -0.026 |
|---|
| S100A1 |   | 2 | 27 | 148 | 148 | -0.115 | 0.023 | 0.050 | 0.108 | 0.039 |
|---|
| HSPB8 |   | 3 | 13 | 1 | 5 | 0.231 | 0.010 | 0.076 | -0.044 | 0.013 |
|---|
| CACNA1S |   | 1 | 59 | 148 | 150 | -0.091 | 0.093 | 0.060 | 0.238 | 0.031 |
|---|
| HSPB1 |   | 6 | 2 | 1 | 1 | 0.262 | 0.053 | 0.231 | 0.043 | 0.144 |
|---|
| STAT3 |   | 2 | 27 | 110 | 113 | 0.100 | -0.078 | -0.046 | 0.077 | -0.189 |
|---|
| SSBP4 |   | 1 | 59 | 148 | 150 | undef | -0.045 | 0.095 | undef | undef |
|---|
| MDFI |   | 1 | 59 | 148 | 150 | -0.021 | -0.109 | -0.069 | 0.271 | 0.105 |
|---|
GO Enrichment output for subnetwork 82 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| pituitary gland development | GO:0021983 |  | 3.126E-06 | 7.19E-03 |
|---|
| diencephalon development | GO:0021536 |  | 4.795E-06 | 5.515E-03 |
|---|
| cell fate determination | GO:0001709 |  | 1.501E-05 | 0.01150864 |
|---|
| response to heat | GO:0009408 |  | 3.406E-05 | 0.01958662 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 5.758E-05 | 0.02648755 |
|---|
| anterograde axon cargo transport | GO:0008089 |  | 6.503E-05 | 0.02492842 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 6.503E-05 | 0.02136721 |
|---|
| cell maturation | GO:0048469 |  | 1.165E-04 | 0.03348822 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 1.211E-04 | 0.03093931 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 1.211E-04 | 0.02784538 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 1.211E-04 | 0.02531398 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anterograde axon cargo transport | GO:0008089 |  | 1.675E-07 | 4.092E-04 |
|---|
| pituitary gland development | GO:0021983 |  | 2.651E-06 | 3.238E-03 |
|---|
| axon cargo transport | GO:0008088 |  | 2.651E-06 | 2.158E-03 |
|---|
| diencephalon development | GO:0021536 |  | 6.257E-06 | 3.821E-03 |
|---|
| microtubule-based transport | GO:0010970 |  | 7.236E-06 | 3.535E-03 |
|---|
| cell fate determination | GO:0001709 |  | 1.889E-05 | 7.692E-03 |
|---|
| response to heat | GO:0009408 |  | 2.089E-05 | 7.29E-03 |
|---|
| neuron fate determination | GO:0048664 |  | 2.887E-05 | 8.818E-03 |
|---|
| phospholipid scrambling | GO:0017121 |  | 2.887E-05 | 7.838E-03 |
|---|
| positive regulation of actin filament polymerization | GO:0030838 |  | 4.326E-05 | 0.01056945 |
|---|
| cytoskeleton-dependent intracellular transport | GO:0030705 |  | 5.652E-05 | 0.01255168 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anterograde axon cargo transport | GO:0008089 |  | 1.792E-07 | 4.312E-04 |
|---|
| pituitary gland development | GO:0021983 |  | 2.307E-06 | 2.775E-03 |
|---|
| axon cargo transport | GO:0008088 |  | 2.835E-06 | 2.274E-03 |
|---|
| diencephalon development | GO:0021536 |  | 5.743E-06 | 3.455E-03 |
|---|
| microtubule-based transport | GO:0010970 |  | 7.739E-06 | 3.724E-03 |
|---|
| cell fate determination | GO:0001709 |  | 1.82E-05 | 7.3E-03 |
|---|
| response to heat | GO:0009408 |  | 2.02E-05 | 6.944E-03 |
|---|
| neuron fate determination | GO:0048664 |  | 3.028E-05 | 9.108E-03 |
|---|
| phospholipid scrambling | GO:0017121 |  | 3.028E-05 | 8.096E-03 |
|---|
| positive regulation of actin filament polymerization | GO:0030838 |  | 4.537E-05 | 0.01091679 |
|---|
| cytoskeleton-dependent intracellular transport | GO:0030705 |  | 5.628E-05 | 0.01230893 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| pituitary gland development | GO:0021983 |  | 3.126E-06 | 7.19E-03 |
|---|
| diencephalon development | GO:0021536 |  | 4.795E-06 | 5.515E-03 |
|---|
| cell fate determination | GO:0001709 |  | 1.501E-05 | 0.01150864 |
|---|
| response to heat | GO:0009408 |  | 3.406E-05 | 0.01958662 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 5.758E-05 | 0.02648755 |
|---|
| anterograde axon cargo transport | GO:0008089 |  | 6.503E-05 | 0.02492842 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 6.503E-05 | 0.02136721 |
|---|
| cell maturation | GO:0048469 |  | 1.165E-04 | 0.03348822 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 1.211E-04 | 0.03093931 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 1.211E-04 | 0.02784538 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 1.211E-04 | 0.02531398 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| pituitary gland development | GO:0021983 |  | 3.126E-06 | 7.19E-03 |
|---|
| diencephalon development | GO:0021536 |  | 4.795E-06 | 5.515E-03 |
|---|
| cell fate determination | GO:0001709 |  | 1.501E-05 | 0.01150864 |
|---|
| response to heat | GO:0009408 |  | 3.406E-05 | 0.01958662 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 5.758E-05 | 0.02648755 |
|---|
| anterograde axon cargo transport | GO:0008089 |  | 6.503E-05 | 0.02492842 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 6.503E-05 | 0.02136721 |
|---|
| cell maturation | GO:0048469 |  | 1.165E-04 | 0.03348822 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 1.211E-04 | 0.03093931 |
|---|
| regulation of Notch signaling pathway | GO:0008593 |  | 1.211E-04 | 0.02784538 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 1.211E-04 | 0.02531398 |
|---|


