Study VanDeVijver-ER-pos
Study informations
14 subnetworks in total page | file
272 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 133
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.5248 | 0.000e+00 | 0.000e+00 | 3.140e-04 | 0.000e+00 |
|---|
| IPC | 0.5956 | 1.712e-02 | 8.997e-03 | 9.363e-01 | 1.442e-04 |
|---|
| Loi | 0.4876 | 0.000e+00 | 1.000e-06 | 4.123e-03 | 0.000e+00 |
|---|
| Schmidt | 0.2890 | 1.605e-02 | 4.659e-03 | 3.162e-01 | 2.364e-05 |
|---|
| Wang | 0.3414 | 1.070e-04 | 2.000e-06 | 4.501e-02 | 9.633e-12 |
|---|
Expression data for subnetwork 133 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
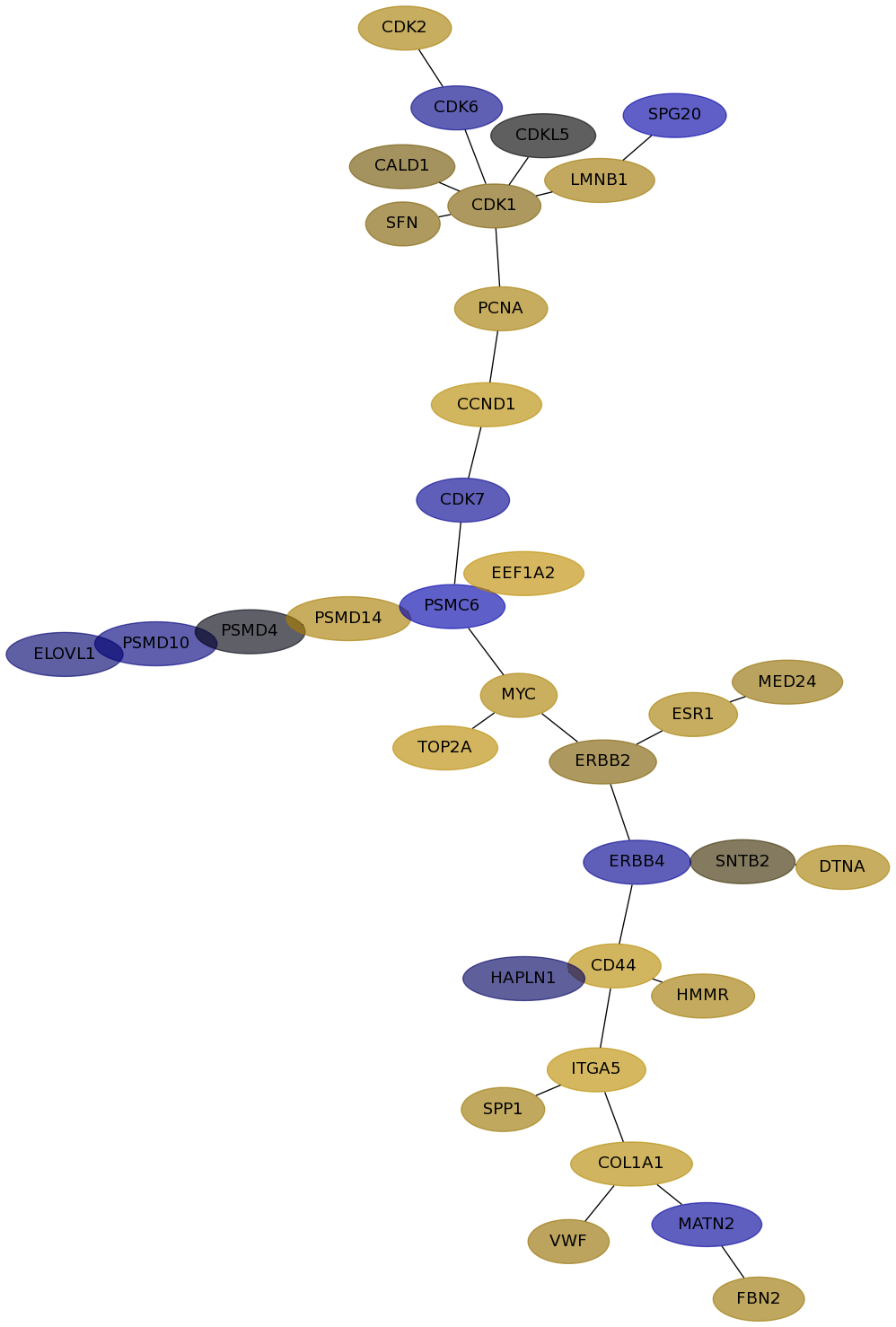
Score for each gene in subnetwork 133 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| PSMD10 |   | 1 | 59 | 36 | 49 | -0.050 | 0.044 | 0.167 | 0.076 | 0.080 |
|---|
| ERBB2 |   | 2 | 27 | 1 | 10 | 0.047 | -0.012 | 0.088 | 0.191 | -0.068 |
|---|
| ELOVL1 |   | 1 | 59 | 36 | 49 | -0.033 | -0.076 | 0.126 | 0.070 | 0.123 |
|---|
| ERBB4 |   | 2 | 27 | 36 | 41 | -0.075 | 0.029 | -0.029 | 0.104 | 0.004 |
|---|
| HAPLN1 |   | 2 | 27 | 36 | 41 | -0.025 | 0.142 | 0.086 | -0.085 | 0.083 |
|---|
| TOP2A |   | 1 | 59 | 36 | 49 | 0.195 | 0.161 | 0.190 | 0.277 | 0.216 |
|---|
| CCND1 |   | 4 | 6 | 36 | 36 | 0.189 | 0.059 | 0.145 | 0.009 | 0.146 |
|---|
| SPP1 |   | 3 | 13 | 36 | 37 | 0.096 | 0.208 | 0.125 | -0.007 | 0.149 |
|---|
| PCNA |   | 3 | 13 | 1 | 5 | 0.120 | 0.192 | 0.141 | 0.138 | 0.184 |
|---|
| LMNB1 |   | 2 | 27 | 36 | 41 | 0.105 | 0.084 | 0.121 | 0.186 | 0.087 |
|---|
| CDKL5 |   | 1 | 59 | 36 | 49 | 0.000 | 0.062 | -0.036 | 0.000 | 0.000 |
|---|
| CALD1 |   | 1 | 59 | 36 | 49 | 0.034 | -0.001 | -0.154 | -0.113 | 0.136 |
|---|
| MYC |   | 4 | 6 | 1 | 3 | 0.142 | 0.031 | 0.041 | 0.035 | -0.138 |
|---|
| EEF1A2 |   | 2 | 27 | 36 | 41 | 0.214 | -0.012 | 0.169 | 0.135 | 0.265 |
|---|
| PSMD14 |   | 2 | 27 | 36 | 41 | 0.127 | 0.104 | -0.014 | -0.129 | 0.224 |
|---|
| MED24 |   | 1 | 59 | 36 | 49 | 0.078 | -0.005 | 0.099 | 0.264 | 0.015 |
|---|
| CD44 |   | 3 | 13 | 36 | 37 | 0.186 | 0.054 | -0.053 | -0.293 | -0.279 |
|---|
| CDK6 |   | 1 | 59 | 36 | 49 | -0.061 | 0.014 | -0.033 | 0.052 | 0.082 |
|---|
| HMMR |   | 2 | 27 | 36 | 41 | 0.108 | 0.202 | 0.167 | 0.194 | 0.282 |
|---|
| DTNA |   | 1 | 59 | 36 | 49 | 0.125 | 0.057 | 0.042 | 0.020 | 0.213 |
|---|
| COL1A1 |   | 1 | 59 | 36 | 49 | 0.174 | -0.136 | -0.017 | -0.082 | 0.097 |
|---|
| SNTB2 |   | 1 | 59 | 36 | 49 | 0.009 | -0.073 | 0.018 | 0.196 | 0.011 |
|---|
| ITGA5 |   | 3 | 13 | 36 | 37 | 0.204 | -0.033 | -0.023 | -0.155 | 0.135 |
|---|
| FBN2 |   | 1 | 59 | 36 | 49 | 0.092 | 0.212 | 0.093 | -0.058 | 0.247 |
|---|
| CDK7 |   | 1 | 59 | 36 | 49 | -0.074 | 0.043 | 0.231 | -0.046 | 0.027 |
|---|
| PSMD4 |   | 1 | 59 | 36 | 49 | -0.002 | 0.140 | 0.156 | 0.081 | 0.194 |
|---|
| CDK1 |   | 3 | 13 | 1 | 5 | 0.047 | 0.152 | 0.200 | 0.204 | 0.254 |
|---|
| PSMC6 |   | 2 | 27 | 36 | 41 | -0.140 | 0.146 | -0.021 | 0.167 | 0.233 |
|---|
| MATN2 |   | 1 | 59 | 36 | 49 | -0.096 | -0.140 | 0.041 | 0.089 | -0.049 |
|---|
| VWF |   | 3 | 13 | 36 | 37 | 0.081 | -0.060 | -0.038 | -0.112 | -0.069 |
|---|
| SPG20 |   | 2 | 27 | 36 | 41 | -0.125 | -0.029 | 0.009 | -0.154 | 0.042 |
|---|
| CDK2 |   | 1 | 59 | 36 | 49 | 0.123 | 0.121 | 0.177 | 0.126 | 0.187 |
|---|
| ESR1 |   | 6 | 2 | 1 | 1 | 0.125 | 0.032 | 0.114 | -0.064 | 0.048 |
|---|
| SFN |   | 1 | 59 | 36 | 49 | 0.048 | 0.043 | 0.107 | 0.083 | 0.056 |
|---|
GO Enrichment output for subnetwork 133 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.088E-10 | 2.503E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.246E-10 | 1.433E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.842E-10 | 1.412E-07 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.842E-10 | 1.059E-07 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 3.013E-10 | 1.386E-07 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 3.81E-10 | 1.46E-07 |
|---|
| regulation of ligase activity | GO:0051340 |  | 5.966E-10 | 1.96E-07 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.009E-09 | 2.901E-07 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 3.96E-09 | 1.012E-06 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 4.459E-09 | 1.026E-06 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 6.113E-07 | 1.278E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.822E-11 | 6.894E-08 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.184E-11 | 3.889E-08 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.525E-11 | 3.685E-08 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.525E-11 | 2.764E-08 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 7.859E-11 | 3.84E-08 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 9.697E-11 | 3.948E-08 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.601E-10 | 5.587E-08 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 3.659E-10 | 1.117E-07 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 1.205E-09 | 3.27E-07 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 1.34E-09 | 3.273E-07 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 2.494E-07 | 5.539E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 5.913E-11 | 1.423E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.67E-11 | 8.024E-08 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 9.472E-11 | 7.597E-08 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 9.472E-11 | 5.698E-08 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.643E-10 | 7.908E-08 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.027E-10 | 8.127E-08 |
|---|
| regulation of ligase activity | GO:0051340 |  | 3.342E-10 | 1.149E-07 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 7.622E-10 | 2.292E-07 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.303E-09 | 6.158E-07 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 2.561E-09 | 6.162E-07 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 3.325E-07 | 7.273E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.088E-10 | 2.503E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.246E-10 | 1.433E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.842E-10 | 1.412E-07 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.842E-10 | 1.059E-07 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 3.013E-10 | 1.386E-07 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 3.81E-10 | 1.46E-07 |
|---|
| regulation of ligase activity | GO:0051340 |  | 5.966E-10 | 1.96E-07 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.009E-09 | 2.901E-07 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 3.96E-09 | 1.012E-06 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 4.459E-09 | 1.026E-06 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 6.113E-07 | 1.278E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.088E-10 | 2.503E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.246E-10 | 1.433E-07 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.842E-10 | 1.412E-07 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.842E-10 | 1.059E-07 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 3.013E-10 | 1.386E-07 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 3.81E-10 | 1.46E-07 |
|---|
| regulation of ligase activity | GO:0051340 |  | 5.966E-10 | 1.96E-07 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.009E-09 | 2.901E-07 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 3.96E-09 | 1.012E-06 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 4.459E-09 | 1.026E-06 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 6.113E-07 | 1.278E-04 |
|---|


