Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 929
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3393 | 1.146e-03 | 1.700e-05 | 1.441e-01 | 2.807e-09 |
|---|
| IPC | 0.3526 | 3.294e-01 | 1.618e-01 | 9.316e-01 | 4.966e-02 |
|---|
| Loi | 0.4497 | 1.830e-04 | 0.000e+00 | 6.817e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5810 | 2.000e-06 | 0.000e+00 | 1.564e-01 | 0.000e+00 |
|---|
| Wang | 0.2517 | 9.551e-03 | 7.620e-02 | 2.546e-01 | 1.853e-04 |
|---|
Expression data for subnetwork 929 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
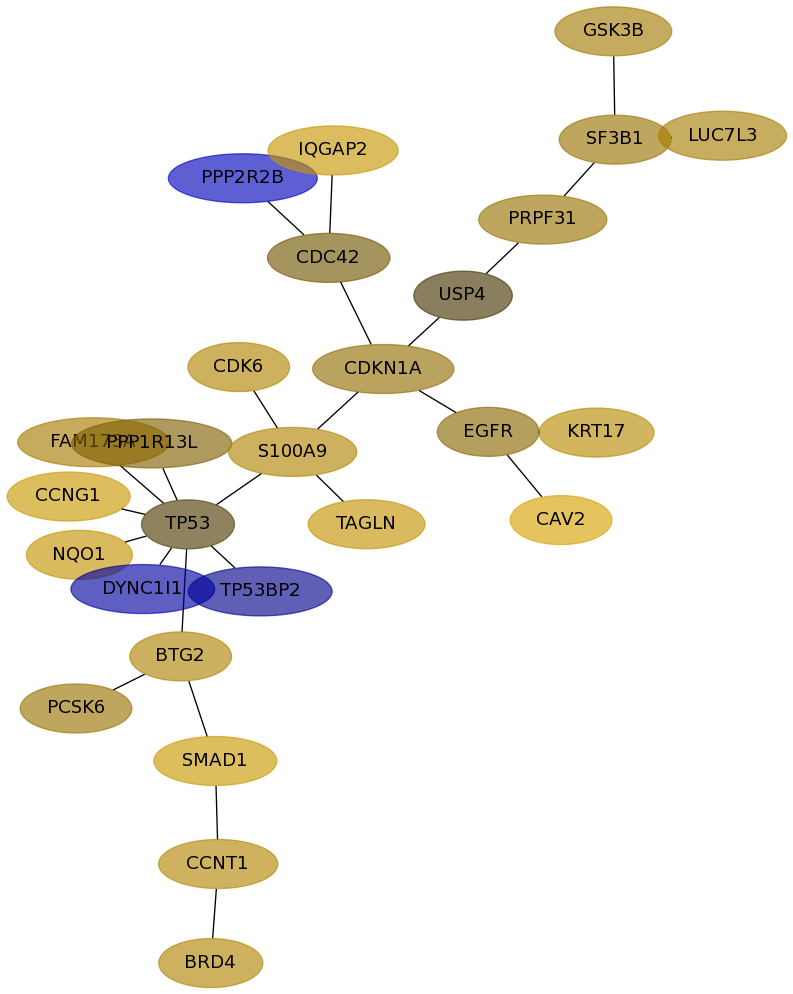
Score for each gene in subnetwork 929 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| KRT17 |   | 2 | 391 | 398 | 411 | 0.187 | -0.129 | 0.174 | -0.183 | -0.072 |
|---|
| USP4 |   | 6 | 120 | 823 | 803 | 0.012 | -0.093 | 0.147 | 0.097 | 0.054 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| NQO1 |   | 6 | 120 | 536 | 535 | 0.243 | -0.115 | 0.069 | -0.189 | 0.118 |
|---|
| PRPF31 |   | 1 | 652 | 1415 | 1415 | 0.089 | 0.167 | 0.076 | 0.119 | -0.089 |
|---|
| BTG2 |   | 6 | 120 | 1 | 21 | 0.146 | -0.029 | -0.093 | -0.111 | -0.170 |
|---|
| SF3B1 |   | 15 | 40 | 148 | 144 | 0.091 | -0.154 | 0.126 | -0.073 | -0.204 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| CCNG1 |   | 4 | 198 | 816 | 802 | 0.287 | -0.100 | 0.061 | -0.157 | -0.093 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| DYNC1I1 |   | 5 | 148 | 182 | 181 | -0.109 | -0.069 | 0.013 | -0.086 | 0.088 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| FAM173A |   | 3 | 265 | 1325 | 1319 | 0.118 | 0.125 | -0.114 | -0.218 | 0.169 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| LUC7L3 |   | 13 | 47 | 148 | 148 | 0.123 | 0.043 | 0.237 | 0.175 | 0.074 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| PPP1R13L |   | 2 | 391 | 283 | 301 | 0.049 | 0.032 | 0.351 | -0.104 | 0.173 |
|---|
| TP53BP2 |   | 5 | 148 | 1 | 22 | -0.067 | 0.245 | -0.012 | 0.129 | 0.017 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| PCSK6 |   | 1 | 652 | 1415 | 1415 | 0.089 | -0.017 | 0.107 | 0.217 | -0.035 |
|---|
| TAGLN |   | 4 | 198 | 823 | 807 | 0.243 | -0.079 | 0.246 | 0.170 | 0.175 |
|---|
GO Enrichment output for subnetwork 929 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| intermediate filament cytoskeleton organization | GO:0045104 |  | 6.607E-08 | 1.52E-04 |
|---|
| intermediate filament-based process | GO:0045103 |  | 9.228E-08 | 1.061E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.099E-07 | 2.376E-04 |
|---|
| nerve growth factor processing | GO:0032455 |  | 3.099E-07 | 1.782E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.099E-07 | 1.426E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.899E-07 | 2.261E-04 |
|---|
| ER overload response | GO:0006983 |  | 6.185E-07 | 2.032E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.185E-07 | 1.778E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 6.616E-07 | 1.691E-04 |
|---|
| embryonic pattern specification | GO:0009880 |  | 7.99E-07 | 1.838E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.08E-06 | 2.258E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| intermediate filament cytoskeleton organization | GO:0045104 |  | 5.135E-08 | 1.254E-04 |
|---|
| intermediate filament-based process | GO:0045103 |  | 6.833E-08 | 8.347E-05 |
|---|
| nerve growth factor processing | GO:0032455 |  | 1.564E-07 | 1.274E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.122E-07 | 1.907E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.122E-07 | 1.526E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.51E-07 | 1.429E-04 |
|---|
| embryonic pattern specification | GO:0009880 |  | 6.45E-07 | 2.251E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 7.511E-07 | 2.294E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 8.711E-07 | 2.364E-04 |
|---|
| ER overload response | GO:0006983 |  | 8.711E-07 | 2.128E-04 |
|---|
| intermediate filament organization | GO:0045109 |  | 8.711E-07 | 1.935E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| intermediate filament cytoskeleton organization | GO:0045104 |  | 6.809E-08 | 1.638E-04 |
|---|
| intermediate filament-based process | GO:0045103 |  | 9.06E-08 | 1.09E-04 |
|---|
| nerve growth factor processing | GO:0032455 |  | 1.938E-07 | 1.554E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.868E-07 | 2.326E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.868E-07 | 1.861E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.954E-07 | 1.987E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.755E-07 | 2.322E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.755E-07 | 2.032E-04 |
|---|
| embryonic pattern specification | GO:0009880 |  | 7.289E-07 | 1.949E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 8.539E-07 | 2.055E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.079E-06 | 2.359E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| intermediate filament cytoskeleton organization | GO:0045104 |  | 6.607E-08 | 1.52E-04 |
|---|
| intermediate filament-based process | GO:0045103 |  | 9.228E-08 | 1.061E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.099E-07 | 2.376E-04 |
|---|
| nerve growth factor processing | GO:0032455 |  | 3.099E-07 | 1.782E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.099E-07 | 1.426E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.899E-07 | 2.261E-04 |
|---|
| ER overload response | GO:0006983 |  | 6.185E-07 | 2.032E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.185E-07 | 1.778E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 6.616E-07 | 1.691E-04 |
|---|
| embryonic pattern specification | GO:0009880 |  | 7.99E-07 | 1.838E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.08E-06 | 2.258E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| intermediate filament cytoskeleton organization | GO:0045104 |  | 6.607E-08 | 1.52E-04 |
|---|
| intermediate filament-based process | GO:0045103 |  | 9.228E-08 | 1.061E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.099E-07 | 2.376E-04 |
|---|
| nerve growth factor processing | GO:0032455 |  | 3.099E-07 | 1.782E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.099E-07 | 1.426E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.899E-07 | 2.261E-04 |
|---|
| ER overload response | GO:0006983 |  | 6.185E-07 | 2.032E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.185E-07 | 1.778E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 6.616E-07 | 1.691E-04 |
|---|
| embryonic pattern specification | GO:0009880 |  | 7.99E-07 | 1.838E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.08E-06 | 2.258E-04 |
|---|


