Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 879
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3489 | 7.290e-04 | 8.000e-06 | 1.181e-01 | 6.885e-10 |
|---|
| IPC | 0.3362 | 3.564e-01 | 1.852e-01 | 9.372e-01 | 6.185e-02 |
|---|
| Loi | 0.4431 | 2.430e-04 | 0.000e+00 | 7.226e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5689 | 5.000e-06 | 0.000e+00 | 1.891e-01 | 0.000e+00 |
|---|
| Wang | 0.2468 | 1.155e-02 | 8.577e-02 | 2.774e-01 | 2.749e-04 |
|---|
Expression data for subnetwork 879 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
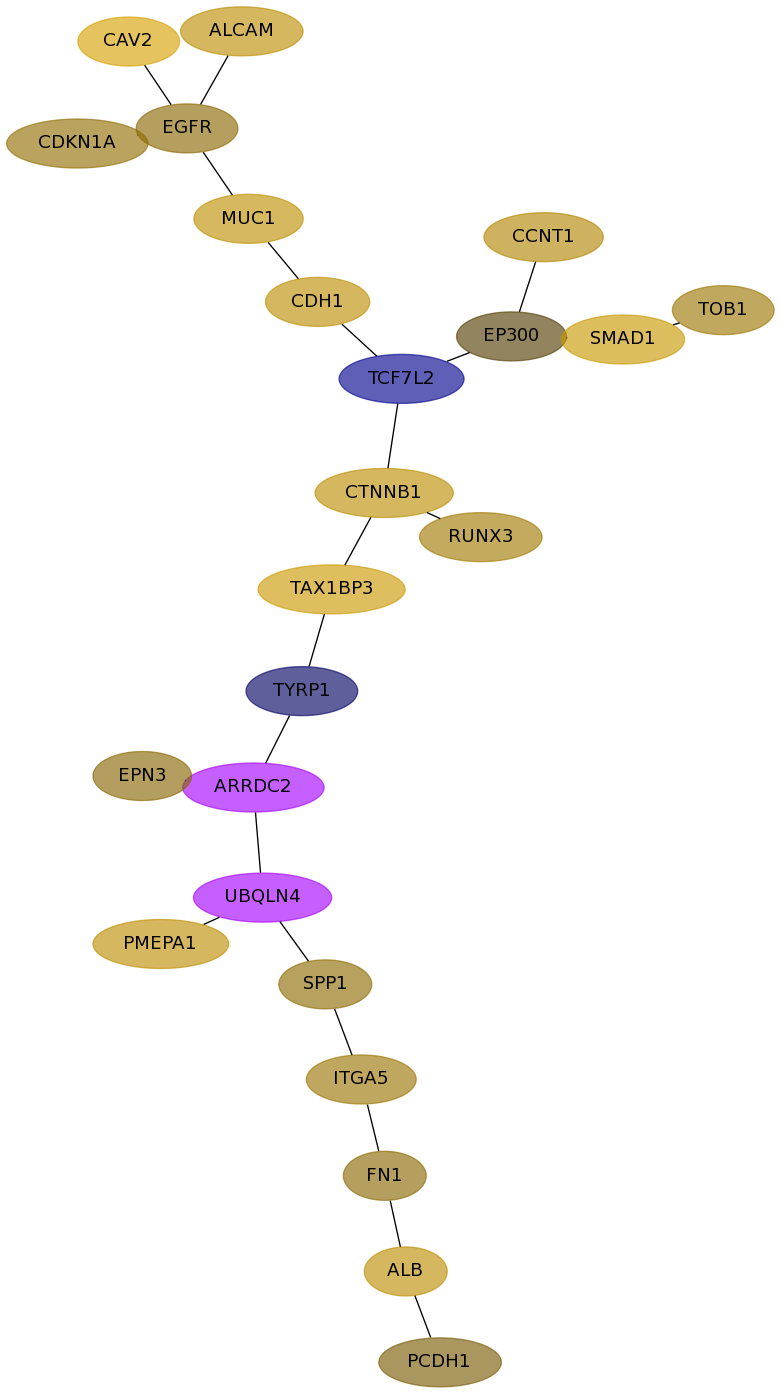
Score for each gene in subnetwork 879 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| UBQLN4 |   | 2 | 391 | 937 | 928 | undef | 0.143 | -0.114 | undef | undef |
|---|
| PCDH1 |   | 2 | 391 | 283 | 301 | 0.042 | 0.104 | 0.266 | -0.067 | 0.001 |
|---|
| FN1 |   | 5 | 148 | 57 | 63 | 0.064 | 0.190 | 0.133 | 0.091 | 0.083 |
|---|
| ITGA5 |   | 5 | 148 | 339 | 337 | 0.093 | 0.031 | 0.192 | 0.125 | 0.202 |
|---|
| RUNX3 |   | 8 | 84 | 33 | 35 | 0.109 | 0.071 | -0.097 | 0.083 | -0.074 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| ALCAM |   | 4 | 198 | 1276 | 1276 | 0.206 | -0.208 | 0.193 | 0.067 | -0.010 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| EPN3 |   | 2 | 391 | 937 | 928 | 0.061 | 0.185 | 0.179 | 0.001 | 0.081 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| SPP1 |   | 4 | 198 | 339 | 341 | 0.068 | 0.130 | 0.119 | 0.184 | 0.153 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| PMEPA1 |   | 4 | 198 | 937 | 910 | 0.208 | -0.085 | 0.137 | 0.167 | 0.102 |
|---|
| ALB |   | 7 | 100 | 57 | 60 | 0.207 | -0.122 | 0.218 | 0.080 | 0.038 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| TAX1BP3 |   | 5 | 148 | 552 | 541 | 0.292 | 0.150 | 0.219 | 0.070 | 0.206 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| ARRDC2 |   | 2 | 391 | 937 | 928 | undef | 0.192 | -0.100 | undef | undef |
|---|
| TOB1 |   | 1 | 652 | 1445 | 1445 | 0.096 | 0.055 | 0.097 | -0.069 | -0.081 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| TYRP1 |   | 1 | 652 | 1445 | 1445 | -0.025 | 0.135 | 0.051 | -0.020 | -0.018 |
|---|
GO Enrichment output for subnetwork 879 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.704E-07 | 1.312E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.9E-06 | 5.635E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.14E-05 | 8.739E-03 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.328E-05 | 7.635E-03 |
|---|
| regulation of osteoblast differentiation | GO:0045667 |  | 2.011E-05 | 9.252E-03 |
|---|
| ossification | GO:0001503 |  | 3.263E-05 | 0.01250975 |
|---|
| skeletal muscle tissue development | GO:0007519 |  | 4.12E-05 | 0.01353711 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 4.829E-05 | 0.01388318 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 4.829E-05 | 0.01234061 |
|---|
| melanin biosynthetic process from tyrosine | GO:0006583 |  | 4.829E-05 | 0.01110655 |
|---|
| melanosome organization | GO:0032438 |  | 4.829E-05 | 0.01009686 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.006E-12 | 1.956E-08 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.163E-07 | 1.42E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 3.247E-07 | 2.644E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 6.427E-07 | 3.925E-04 |
|---|
| positive regulation of peptide secretion | GO:0002793 |  | 9.53E-07 | 4.657E-04 |
|---|
| skeletal muscle tissue development | GO:0007519 |  | 1.151E-06 | 4.687E-04 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 1.269E-06 | 4.429E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 2.614E-06 | 7.984E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.74E-06 | 1.015E-03 |
|---|
| myoblast differentiation | GO:0045445 |  | 3.897E-06 | 9.52E-04 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 5.539E-06 | 1.23E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.075E-12 | 1.221E-08 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.28E-07 | 1.54E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 3.575E-07 | 2.867E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 6.49E-07 | 3.904E-04 |
|---|
| positive regulation of peptide secretion | GO:0002793 |  | 1.049E-06 | 5.049E-04 |
|---|
| skeletal muscle tissue development | GO:0007519 |  | 1.267E-06 | 5.082E-04 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 1.397E-06 | 4.802E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 2.878E-06 | 8.654E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.225E-06 | 1.13E-03 |
|---|
| myoblast differentiation | GO:0045445 |  | 4.289E-06 | 1.032E-03 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 6.096E-06 | 1.333E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.704E-07 | 1.312E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.9E-06 | 5.635E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.14E-05 | 8.739E-03 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.328E-05 | 7.635E-03 |
|---|
| regulation of osteoblast differentiation | GO:0045667 |  | 2.011E-05 | 9.252E-03 |
|---|
| ossification | GO:0001503 |  | 3.263E-05 | 0.01250975 |
|---|
| skeletal muscle tissue development | GO:0007519 |  | 4.12E-05 | 0.01353711 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 4.829E-05 | 0.01388318 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 4.829E-05 | 0.01234061 |
|---|
| melanin biosynthetic process from tyrosine | GO:0006583 |  | 4.829E-05 | 0.01110655 |
|---|
| melanosome organization | GO:0032438 |  | 4.829E-05 | 0.01009686 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.704E-07 | 1.312E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.9E-06 | 5.635E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.14E-05 | 8.739E-03 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.328E-05 | 7.635E-03 |
|---|
| regulation of osteoblast differentiation | GO:0045667 |  | 2.011E-05 | 9.252E-03 |
|---|
| ossification | GO:0001503 |  | 3.263E-05 | 0.01250975 |
|---|
| skeletal muscle tissue development | GO:0007519 |  | 4.12E-05 | 0.01353711 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 4.829E-05 | 0.01388318 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 4.829E-05 | 0.01234061 |
|---|
| melanin biosynthetic process from tyrosine | GO:0006583 |  | 4.829E-05 | 0.01110655 |
|---|
| melanosome organization | GO:0032438 |  | 4.829E-05 | 0.01009686 |
|---|


