Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 752
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3267 | 2.028e-03 | 4.300e-05 | 1.843e-01 | 1.607e-08 |
|---|
| IPC | 0.3527 | 3.293e-01 | 1.617e-01 | 9.316e-01 | 4.960e-02 |
|---|
| Loi | 0.4467 | 2.090e-04 | 0.000e+00 | 7.003e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5833 | 2.000e-06 | 0.000e+00 | 1.506e-01 | 0.000e+00 |
|---|
| Wang | 0.2389 | 1.544e-02 | 1.026e-01 | 3.152e-01 | 4.995e-04 |
|---|
Expression data for subnetwork 752 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
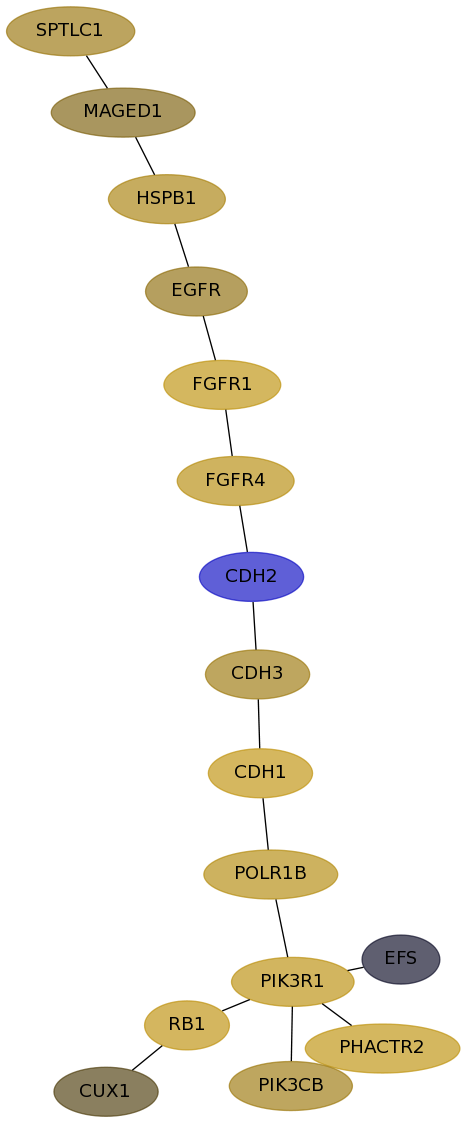
Score for each gene in subnetwork 752 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| MAGED1 |   | 6 | 120 | 715 | 695 | 0.040 | -0.141 | 0.284 | -0.152 | 0.045 |
|---|
| CUX1 |   | 8 | 84 | 271 | 266 | 0.012 | -0.042 | 0.256 | -0.121 | 0.040 |
|---|
| PHACTR2 |   | 5 | 148 | 271 | 267 | 0.203 | -0.057 | -0.051 | 0.143 | 0.164 |
|---|
| EFS |   | 4 | 198 | 129 | 130 | -0.003 | -0.024 | 0.300 | -0.003 | -0.034 |
|---|
| PIK3CB |   | 2 | 391 | 1188 | 1174 | 0.088 | 0.111 | 0.108 | -0.158 | 0.068 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CDH3 |   | 10 | 67 | 1 | 16 | 0.088 | 0.028 | 0.169 | -0.055 | -0.027 |
|---|
| POLR1B |   | 11 | 59 | 129 | 126 | 0.157 | 0.062 | -0.036 | -0.079 | 0.139 |
|---|
| SPTLC1 |   | 1 | 652 | 1442 | 1442 | 0.081 | -0.093 | 0.049 | 0.018 | 0.191 |
|---|
| HSPB1 |   | 4 | 198 | 148 | 162 | 0.121 | 0.053 | 0.169 | 0.017 | 0.161 |
|---|
| CDH2 |   | 7 | 100 | 33 | 37 | -0.228 | 0.169 | 0.109 | -0.016 | 0.155 |
|---|
GO Enrichment output for subnetwork 752 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide phosphorylation | GO:0046854 |  | 1.494E-08 | 3.435E-05 |
|---|
| lipid phosphorylation | GO:0046834 |  | 2.034E-08 | 2.339E-05 |
|---|
| homophilic cell adhesion | GO:0007156 |  | 5.103E-08 | 3.912E-05 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 1.742E-06 | 1.002E-03 |
|---|
| lipid modification | GO:0030258 |  | 3.26E-06 | 1.499E-03 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 4.409E-06 | 1.69E-03 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 5.346E-06 | 1.757E-03 |
|---|
| lung development | GO:0030324 |  | 6.357E-06 | 1.828E-03 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 6.406E-06 | 1.637E-03 |
|---|
| phosphoinositide metabolic process | GO:0030384 |  | 6.802E-06 | 1.564E-03 |
|---|
| respiratory tube development | GO:0030323 |  | 7.758E-06 | 1.622E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 5.91E-11 | 1.444E-07 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 2.5E-10 | 3.053E-07 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 2.5E-10 | 2.036E-07 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 4.024E-10 | 2.458E-07 |
|---|
| lung development | GO:0030324 |  | 1.601E-09 | 7.825E-07 |
|---|
| respiratory tube development | GO:0030323 |  | 2.079E-09 | 8.466E-07 |
|---|
| respiratory system development | GO:0060541 |  | 2.459E-09 | 8.583E-07 |
|---|
| inner ear development | GO:0048839 |  | 2.621E-08 | 8.004E-06 |
|---|
| phosphoinositide phosphorylation | GO:0046854 |  | 3.032E-08 | 8.23E-06 |
|---|
| lipid phosphorylation | GO:0046834 |  | 3.741E-08 | 9.138E-06 |
|---|
| embryonic cleavage | GO:0040016 |  | 4.114E-08 | 9.136E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 4.902E-11 | 1.179E-07 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 1.731E-10 | 2.082E-07 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 2.269E-10 | 1.82E-07 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 2.933E-10 | 1.764E-07 |
|---|
| lung development | GO:0030324 |  | 1.604E-09 | 7.721E-07 |
|---|
| respiratory tube development | GO:0030323 |  | 2.1E-09 | 8.421E-07 |
|---|
| respiratory system development | GO:0060541 |  | 2.496E-09 | 8.579E-07 |
|---|
| phosphoinositide phosphorylation | GO:0046854 |  | 2.193E-08 | 6.594E-06 |
|---|
| inner ear development | GO:0048839 |  | 2.277E-08 | 6.087E-06 |
|---|
| lipid phosphorylation | GO:0046834 |  | 2.774E-08 | 6.674E-06 |
|---|
| embryonic cleavage | GO:0040016 |  | 4.564E-08 | 9.982E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide phosphorylation | GO:0046854 |  | 1.494E-08 | 3.435E-05 |
|---|
| lipid phosphorylation | GO:0046834 |  | 2.034E-08 | 2.339E-05 |
|---|
| homophilic cell adhesion | GO:0007156 |  | 5.103E-08 | 3.912E-05 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 1.742E-06 | 1.002E-03 |
|---|
| lipid modification | GO:0030258 |  | 3.26E-06 | 1.499E-03 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 4.409E-06 | 1.69E-03 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 5.346E-06 | 1.757E-03 |
|---|
| lung development | GO:0030324 |  | 6.357E-06 | 1.828E-03 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 6.406E-06 | 1.637E-03 |
|---|
| phosphoinositide metabolic process | GO:0030384 |  | 6.802E-06 | 1.564E-03 |
|---|
| respiratory tube development | GO:0030323 |  | 7.758E-06 | 1.622E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide phosphorylation | GO:0046854 |  | 1.494E-08 | 3.435E-05 |
|---|
| lipid phosphorylation | GO:0046834 |  | 2.034E-08 | 2.339E-05 |
|---|
| homophilic cell adhesion | GO:0007156 |  | 5.103E-08 | 3.912E-05 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 1.742E-06 | 1.002E-03 |
|---|
| lipid modification | GO:0030258 |  | 3.26E-06 | 1.499E-03 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 4.409E-06 | 1.69E-03 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 5.346E-06 | 1.757E-03 |
|---|
| lung development | GO:0030324 |  | 6.357E-06 | 1.828E-03 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 6.406E-06 | 1.637E-03 |
|---|
| phosphoinositide metabolic process | GO:0030384 |  | 6.802E-06 | 1.564E-03 |
|---|
| respiratory tube development | GO:0030323 |  | 7.758E-06 | 1.622E-03 |
|---|


