Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 729
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3202 | 2.676e-03 | 6.800e-05 | 2.072e-01 | 3.770e-08 |
|---|
| IPC | 0.3629 | 3.124e-01 | 1.479e-01 | 9.280e-01 | 4.287e-02 |
|---|
| Loi | 0.4451 | 2.240e-04 | 0.000e+00 | 7.102e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5919 | 1.000e-06 | 0.000e+00 | 1.302e-01 | 0.000e+00 |
|---|
| Wang | 0.2393 | 1.521e-02 | 1.017e-01 | 3.132e-01 | 4.842e-04 |
|---|
Expression data for subnetwork 729 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
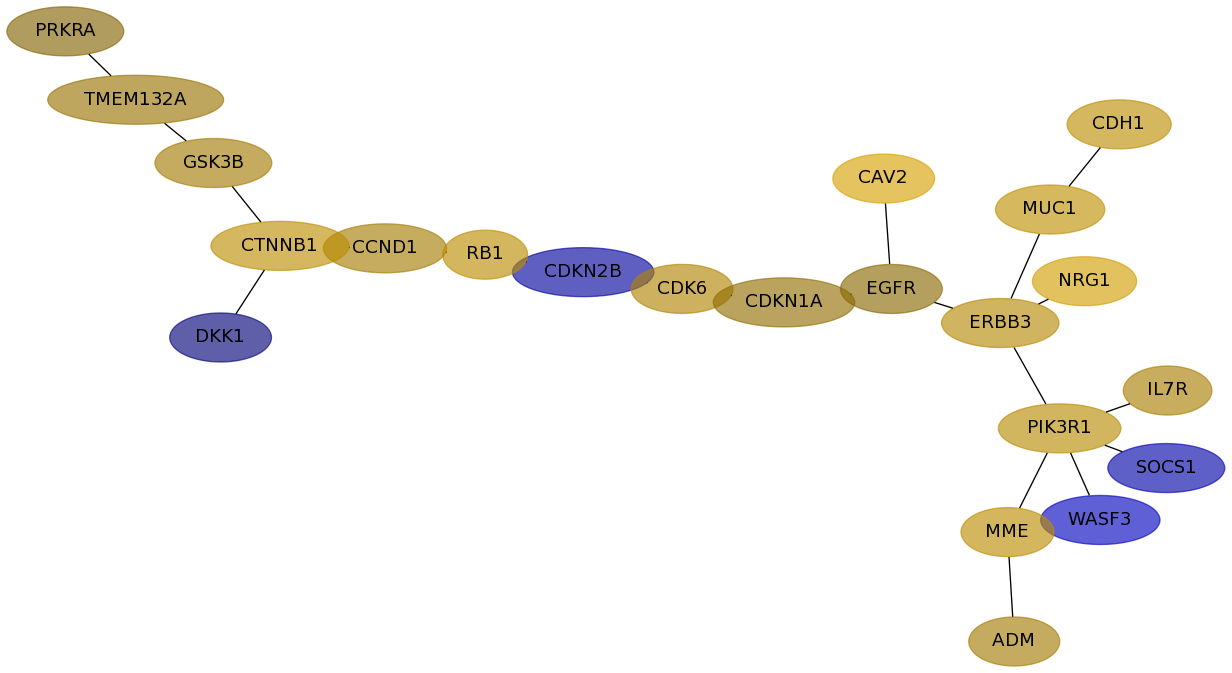
Score for each gene in subnetwork 729 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| CDKN2B |   | 4 | 198 | 937 | 910 | -0.099 | 0.066 | 0.050 | -0.028 | 0.153 |
|---|
| WASF3 |   | 2 | 391 | 1070 | 1062 | -0.213 | 0.038 | -0.040 | 0.036 | 0.054 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| TMEM132A |   | 7 | 100 | 33 | 37 | 0.088 | 0.019 | 0.127 | -0.141 | -0.092 |
|---|
| PRKRA |   | 1 | 652 | 1420 | 1420 | 0.055 | -0.137 | 0.010 | 0.088 | -0.086 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| ADM |   | 2 | 391 | 1188 | 1174 | 0.116 | 0.255 | 0.187 | 0.091 | 0.212 |
|---|
| DKK1 |   | 4 | 198 | 271 | 273 | -0.041 | 0.134 | 0.091 | -0.124 | 0.129 |
|---|
| CCND1 |   | 12 | 55 | 148 | 150 | 0.122 | -0.258 | 0.074 | 0.117 | 0.003 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| SOCS1 |   | 1 | 652 | 1420 | 1420 | -0.132 | 0.074 | -0.153 | -0.177 | -0.100 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| IL7R |   | 14 | 42 | 339 | 334 | 0.130 | -0.151 | -0.112 | -0.130 | -0.160 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| NRG1 |   | 3 | 265 | 262 | 270 | 0.341 | -0.035 | 0.020 | -0.123 | -0.130 |
|---|
| ERBB3 |   | 5 | 148 | 206 | 202 | 0.175 | -0.171 | -0.007 | -0.141 | -0.098 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| MME |   | 8 | 84 | 57 | 54 | 0.200 | -0.121 | 0.020 | 0.088 | -0.130 |
|---|
GO Enrichment output for subnetwork 729 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.798E-09 | 4.136E-06 |
|---|
| interphase | GO:0051325 |  | 2.117E-09 | 2.434E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.162E-08 | 5.491E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.722E-07 | 9.901E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.984E-07 | 1.373E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.247E-07 | 1.628E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.17E-07 | 1.699E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 5.17E-07 | 1.486E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.257E-07 | 2.11E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.692E-06 | 6.191E-04 |
|---|
| response to UV | GO:0009411 |  | 5.272E-06 | 1.102E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.399E-10 | 1.808E-06 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 3.766E-09 | 4.601E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.639E-08 | 2.149E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.933E-08 | 4.845E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.191E-07 | 5.817E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.67E-07 | 6.801E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 2.228E-07 | 7.777E-05 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.561E-07 | 1.087E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 7.265E-07 | 1.972E-04 |
|---|
| mesenchymal cell development | GO:0014031 |  | 1.108E-06 | 2.706E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.62E-06 | 3.599E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.189E-09 | 2.86E-06 |
|---|
| interphase | GO:0051325 |  | 1.481E-09 | 1.782E-06 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 3.616E-09 | 2.9E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.37E-08 | 2.629E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.312E-07 | 6.311E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.79E-07 | 7.177E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.51E-07 | 8.628E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 3.035E-07 | 9.127E-05 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.849E-07 | 1.296E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 9.737E-07 | 2.343E-04 |
|---|
| mesenchymal cell development | GO:0014031 |  | 1.501E-06 | 3.284E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.798E-09 | 4.136E-06 |
|---|
| interphase | GO:0051325 |  | 2.117E-09 | 2.434E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.162E-08 | 5.491E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.722E-07 | 9.901E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.984E-07 | 1.373E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.247E-07 | 1.628E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.17E-07 | 1.699E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 5.17E-07 | 1.486E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.257E-07 | 2.11E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.692E-06 | 6.191E-04 |
|---|
| response to UV | GO:0009411 |  | 5.272E-06 | 1.102E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.798E-09 | 4.136E-06 |
|---|
| interphase | GO:0051325 |  | 2.117E-09 | 2.434E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.162E-08 | 5.491E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.722E-07 | 9.901E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.984E-07 | 1.373E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.247E-07 | 1.628E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.17E-07 | 1.699E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 5.17E-07 | 1.486E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.257E-07 | 2.11E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.692E-06 | 6.191E-04 |
|---|
| response to UV | GO:0009411 |  | 5.272E-06 | 1.102E-03 |
|---|


