Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 643
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2939 | 7.671e-03 | 3.870e-04 | 3.156e-01 | 9.368e-07 |
|---|
| IPC | 0.4082 | 2.401e-01 | 9.468e-02 | 9.120e-01 | 2.073e-02 |
|---|
| Loi | 0.4394 | 2.840e-04 | 0.000e+00 | 7.463e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6228 | 0.000e+00 | 0.000e+00 | 7.271e-02 | 0.000e+00 |
|---|
| Wang | 0.2384 | 1.574e-02 | 1.039e-01 | 3.179e-01 | 5.197e-04 |
|---|
Expression data for subnetwork 643 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
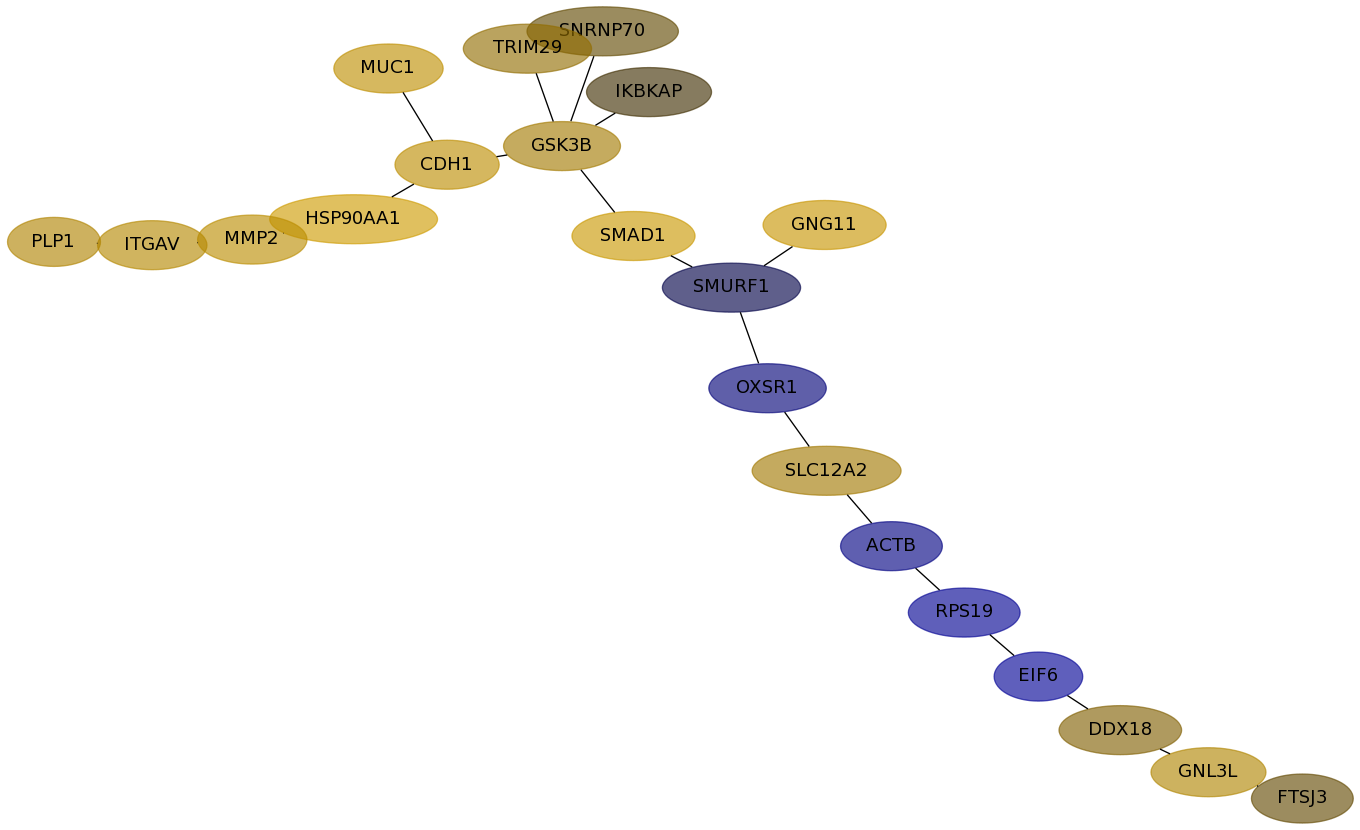
Score for each gene in subnetwork 643 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| GNG11 |   | 4 | 198 | 33 | 41 | 0.270 | 0.049 | -0.039 | 0.135 | 0.106 |
|---|
| DDX18 |   | 1 | 652 | 994 | 1003 | 0.050 | 0.112 | -0.142 | 0.000 | 0.178 |
|---|
| IKBKAP |   | 5 | 148 | 364 | 358 | 0.010 | 0.089 | 0.184 | 0.142 | -0.017 |
|---|
| OXSR1 |   | 2 | 391 | 994 | 983 | -0.040 | -0.109 | 0.199 | 0.223 | -0.072 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| EIF6 |   | 1 | 652 | 994 | 1003 | -0.084 | -0.037 | 0.035 | -0.234 | -0.019 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| MMP2 |   | 7 | 100 | 797 | 778 | 0.207 | -0.105 | 0.186 | -0.024 | 0.162 |
|---|
| RPS19 |   | 1 | 652 | 994 | 1003 | -0.076 | 0.009 | -0.128 | -0.024 | -0.215 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| ACTB |   | 7 | 100 | 398 | 385 | -0.052 | -0.039 | 0.283 | -0.061 | 0.003 |
|---|
| PLP1 |   | 1 | 652 | 994 | 1003 | 0.156 | -0.137 | 0.021 | -0.128 | 0.180 |
|---|
| GNL3L |   | 3 | 265 | 994 | 971 | 0.158 | 0.140 | 0.020 | -0.264 | -0.031 |
|---|
| ITGAV |   | 2 | 391 | 679 | 678 | 0.174 | -0.051 | 0.203 | 0.155 | 0.105 |
|---|
| HSP90AA1 |   | 10 | 67 | 1 | 16 | 0.321 | -0.143 | 0.104 | 0.123 | -0.054 |
|---|
| SNRNP70 |   | 3 | 265 | 994 | 971 | 0.024 | -0.129 | 0.130 | -0.059 | 0.036 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| SMURF1 |   | 9 | 77 | 33 | 34 | -0.013 | -0.222 | 0.236 | -0.049 | -0.006 |
|---|
| SLC12A2 |   | 2 | 391 | 994 | 983 | 0.110 | 0.043 | 0.143 | -0.029 | -0.247 |
|---|
| FTSJ3 |   | 1 | 652 | 994 | 1003 | 0.025 | -0.016 | 0.102 | 0.015 | 0.019 |
|---|
| TRIM29 |   | 2 | 391 | 740 | 745 | 0.077 | 0.161 | 0.240 | -0.150 | 0.024 |
|---|
GO Enrichment output for subnetwork 643 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of transforming growth factor beta receptor signaling pathway | GO:0030512 |  | 4.409E-06 | 0.01014072 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 5.346E-06 | 6.148E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 7.597E-06 | 5.824E-03 |
|---|
| apoptotic cell clearance | GO:0043277 |  | 4.107E-05 | 0.02361515 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 5.016E-05 | 0.02307495 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 6.152E-05 | 0.02358453 |
|---|
| protein refolding | GO:0042026 |  | 6.152E-05 | 0.02021532 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 6.152E-05 | 0.0176884 |
|---|
| hyperosmotic response | GO:0006972 |  | 6.152E-05 | 0.01572302 |
|---|
| ER overload response | GO:0006983 |  | 6.152E-05 | 0.01415072 |
|---|
| protein ubiquitination during ubiquitin-dependent protein catabolic process | GO:0042787 |  | 6.152E-05 | 0.01286429 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gas transport | GO:0015669 |  | 1.362E-06 | 3.327E-03 |
|---|
| negative regulation of transforming growth factor beta receptor signaling pathway | GO:0030512 |  | 1.633E-06 | 1.994E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.277E-06 | 1.854E-03 |
|---|
| ribosomal small subunit biogenesis | GO:0042274 |  | 2.654E-06 | 1.621E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 3.07E-06 | 1.5E-03 |
|---|
| erythrocyte differentiation | GO:0030218 |  | 8.889E-06 | 3.619E-03 |
|---|
| apoptotic cell clearance | GO:0043277 |  | 1.634E-05 | 5.704E-03 |
|---|
| erythrocyte homeostasis | GO:0034101 |  | 1.794E-05 | 5.479E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.248E-05 | 6.101E-03 |
|---|
| protein refolding | GO:0042026 |  | 2.45E-05 | 5.985E-03 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 2.45E-05 | 5.44E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gas transport | GO:0015669 |  | 2.089E-06 | 5.026E-03 |
|---|
| negative regulation of transforming growth factor beta receptor signaling pathway | GO:0030512 |  | 2.504E-06 | 3.012E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 3.491E-06 | 2.8E-03 |
|---|
| ribosomal small subunit biogenesis | GO:0042274 |  | 3.491E-06 | 2.1E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 4.706E-06 | 2.264E-03 |
|---|
| erythrocyte differentiation | GO:0030218 |  | 1.361E-05 | 5.456E-03 |
|---|
| apoptotic cell clearance | GO:0043277 |  | 2.177E-05 | 7.482E-03 |
|---|
| erythrocyte homeostasis | GO:0034101 |  | 2.535E-05 | 7.624E-03 |
|---|
| protein refolding | GO:0042026 |  | 3.262E-05 | 8.721E-03 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 3.262E-05 | 7.849E-03 |
|---|
| hyperosmotic response | GO:0006972 |  | 3.262E-05 | 7.135E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of transforming growth factor beta receptor signaling pathway | GO:0030512 |  | 4.409E-06 | 0.01014072 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 5.346E-06 | 6.148E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 7.597E-06 | 5.824E-03 |
|---|
| apoptotic cell clearance | GO:0043277 |  | 4.107E-05 | 0.02361515 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 5.016E-05 | 0.02307495 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 6.152E-05 | 0.02358453 |
|---|
| protein refolding | GO:0042026 |  | 6.152E-05 | 0.02021532 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 6.152E-05 | 0.0176884 |
|---|
| hyperosmotic response | GO:0006972 |  | 6.152E-05 | 0.01572302 |
|---|
| ER overload response | GO:0006983 |  | 6.152E-05 | 0.01415072 |
|---|
| protein ubiquitination during ubiquitin-dependent protein catabolic process | GO:0042787 |  | 6.152E-05 | 0.01286429 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of transforming growth factor beta receptor signaling pathway | GO:0030512 |  | 4.409E-06 | 0.01014072 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 5.346E-06 | 6.148E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 7.597E-06 | 5.824E-03 |
|---|
| apoptotic cell clearance | GO:0043277 |  | 4.107E-05 | 0.02361515 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 5.016E-05 | 0.02307495 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 6.152E-05 | 0.02358453 |
|---|
| protein refolding | GO:0042026 |  | 6.152E-05 | 0.02021532 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 6.152E-05 | 0.0176884 |
|---|
| hyperosmotic response | GO:0006972 |  | 6.152E-05 | 0.01572302 |
|---|
| ER overload response | GO:0006983 |  | 6.152E-05 | 0.01415072 |
|---|
| protein ubiquitination during ubiquitin-dependent protein catabolic process | GO:0042787 |  | 6.152E-05 | 0.01286429 |
|---|


