Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 589
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2764 | 1.435e-02 | 1.097e-03 | 3.965e-01 | 6.241e-06 |
|---|
| IPC | 0.4372 | 1.972e-01 | 6.801e-02 | 9.017e-01 | 1.209e-02 |
|---|
| Loi | 0.4374 | 3.100e-04 | 0.000e+00 | 7.595e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6378 | 0.000e+00 | 0.000e+00 | 5.285e-02 | 0.000e+00 |
|---|
| Wang | 0.2393 | 1.524e-02 | 1.018e-01 | 3.134e-01 | 4.862e-04 |
|---|
Expression data for subnetwork 589 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
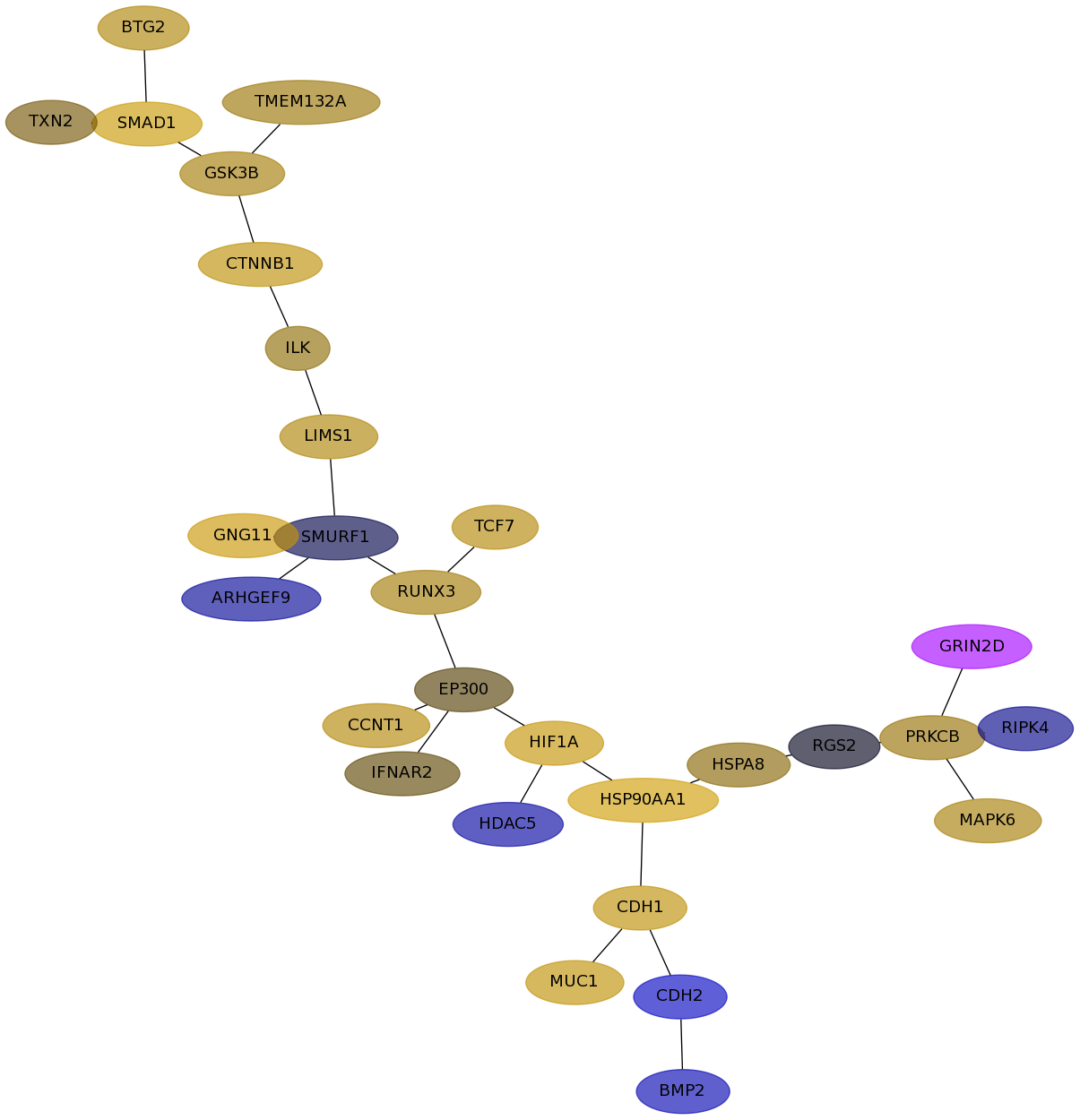
Score for each gene in subnetwork 589 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| IFNAR2 |   | 4 | 198 | 33 | 41 | 0.021 | -0.100 | 0.055 | 0.106 | -0.058 |
|---|
| LIMS1 |   | 2 | 391 | 33 | 55 | 0.145 | 0.063 | 0.090 | -0.032 | 0.277 |
|---|
| GNG11 |   | 4 | 198 | 33 | 41 | 0.270 | 0.049 | -0.039 | 0.135 | 0.106 |
|---|
| BTG2 |   | 6 | 120 | 1 | 21 | 0.146 | -0.029 | -0.093 | -0.111 | -0.170 |
|---|
| HSPA8 |   | 2 | 391 | 33 | 55 | 0.058 | -0.095 | 0.011 | -0.039 | -0.103 |
|---|
| RIPK4 |   | 1 | 652 | 33 | 77 | -0.062 | 0.050 | 0.185 | 0.041 | -0.159 |
|---|
| RUNX3 |   | 8 | 84 | 33 | 35 | 0.109 | 0.071 | -0.097 | 0.083 | -0.074 |
|---|
| TCF7 |   | 1 | 652 | 33 | 77 | 0.164 | 0.102 | 0.054 | 0.048 | 0.123 |
|---|
| ILK |   | 6 | 120 | 33 | 40 | 0.069 | -0.020 | 0.195 | 0.076 | 0.086 |
|---|
| RGS2 |   | 2 | 391 | 33 | 55 | -0.003 | 0.118 | 0.061 | 0.084 | 0.178 |
|---|
| TMEM132A |   | 7 | 100 | 33 | 37 | 0.088 | 0.019 | 0.127 | -0.141 | -0.092 |
|---|
| HDAC5 |   | 2 | 391 | 33 | 55 | -0.107 | 0.022 | 0.029 | -0.165 | 0.009 |
|---|
| BMP2 |   | 7 | 100 | 33 | 37 | -0.155 | -0.011 | 0.097 | 0.148 | -0.069 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| PRKCB |   | 8 | 84 | 33 | 35 | 0.081 | -0.065 | 0.001 | -0.148 | -0.035 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| MAPK6 |   | 1 | 652 | 33 | 77 | 0.119 | -0.174 | 0.213 | -0.137 | 0.155 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| HSP90AA1 |   | 10 | 67 | 1 | 16 | 0.321 | -0.143 | 0.104 | 0.123 | -0.054 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| CDH2 |   | 7 | 100 | 33 | 37 | -0.228 | 0.169 | 0.109 | -0.016 | 0.155 |
|---|
| TXN2 |   | 4 | 198 | 33 | 41 | 0.035 | -0.112 | -0.009 | -0.044 | -0.139 |
|---|
| SMURF1 |   | 9 | 77 | 33 | 34 | -0.013 | -0.222 | 0.236 | -0.049 | -0.006 |
|---|
| ARHGEF9 |   | 2 | 391 | 33 | 55 | -0.084 | -0.051 | 0.134 | 0.173 | 0.132 |
|---|
GO Enrichment output for subnetwork 589 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 4.907E-08 | 1.129E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.077E-06 | 1.239E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 2.011E-06 | 1.542E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.76E-06 | 1.587E-03 |
|---|
| mesenchymal cell development | GO:0014031 |  | 3.447E-06 | 1.586E-03 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 6.923E-06 | 2.654E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.286E-05 | 4.225E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.825E-05 | 5.247E-03 |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 2.143E-05 | 5.477E-03 |
|---|
| cell aging | GO:0007569 |  | 2.495E-05 | 5.74E-03 |
|---|
| ureteric bud development | GO:0001657 |  | 2.884E-05 | 6.03E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 5.175E-11 | 1.264E-07 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 3.328E-09 | 4.065E-06 |
|---|
| mesenchymal cell development | GO:0014031 |  | 1.245E-08 | 1.014E-05 |
|---|
| cell aging | GO:0007569 |  | 7.266E-08 | 4.438E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 6.815E-07 | 3.33E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 9.399E-07 | 3.827E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 2.094E-06 | 7.309E-04 |
|---|
| aging | GO:0007568 |  | 4.011E-06 | 1.225E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 6.508E-06 | 1.766E-03 |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 7.582E-06 | 1.852E-03 |
|---|
| ureteric bud development | GO:0001657 |  | 8.768E-06 | 1.947E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 9.267E-11 | 2.23E-07 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 3.387E-09 | 4.074E-06 |
|---|
| mesenchymal cell development | GO:0014031 |  | 1.952E-08 | 1.566E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 9.064E-07 | 5.452E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 1.273E-06 | 6.125E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 2.975E-06 | 1.193E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 9.237E-06 | 3.175E-03 |
|---|
| cell aging | GO:0007569 |  | 1.076E-05 | 3.236E-03 |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 1.076E-05 | 2.877E-03 |
|---|
| ureteric bud development | GO:0001657 |  | 1.244E-05 | 2.993E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.244E-05 | 2.721E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 4.907E-08 | 1.129E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.077E-06 | 1.239E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 2.011E-06 | 1.542E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.76E-06 | 1.587E-03 |
|---|
| mesenchymal cell development | GO:0014031 |  | 3.447E-06 | 1.586E-03 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 6.923E-06 | 2.654E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.286E-05 | 4.225E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.825E-05 | 5.247E-03 |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 2.143E-05 | 5.477E-03 |
|---|
| cell aging | GO:0007569 |  | 2.495E-05 | 5.74E-03 |
|---|
| ureteric bud development | GO:0001657 |  | 2.884E-05 | 6.03E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 4.907E-08 | 1.129E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.077E-06 | 1.239E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 2.011E-06 | 1.542E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.76E-06 | 1.587E-03 |
|---|
| mesenchymal cell development | GO:0014031 |  | 3.447E-06 | 1.586E-03 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 6.923E-06 | 2.654E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.286E-05 | 4.225E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.825E-05 | 5.247E-03 |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 2.143E-05 | 5.477E-03 |
|---|
| cell aging | GO:0007569 |  | 2.495E-05 | 5.74E-03 |
|---|
| ureteric bud development | GO:0001657 |  | 2.884E-05 | 6.03E-03 |
|---|


