Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1317
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3413 | 1.047e-03 | 1.400e-05 | 1.385e-01 | 2.030e-09 |
|---|
| IPC | 0.3046 | 4.077e-01 | 2.332e-01 | 9.476e-01 | 9.010e-02 |
|---|
| Loi | 0.4909 | 2.700e-05 | 0.000e+00 | 4.642e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5919 | 1.000e-06 | 0.000e+00 | 1.302e-01 | 0.000e+00 |
|---|
| Wang | 0.2523 | 9.323e-03 | 7.506e-02 | 2.518e-01 | 1.762e-04 |
|---|
Expression data for subnetwork 1317 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
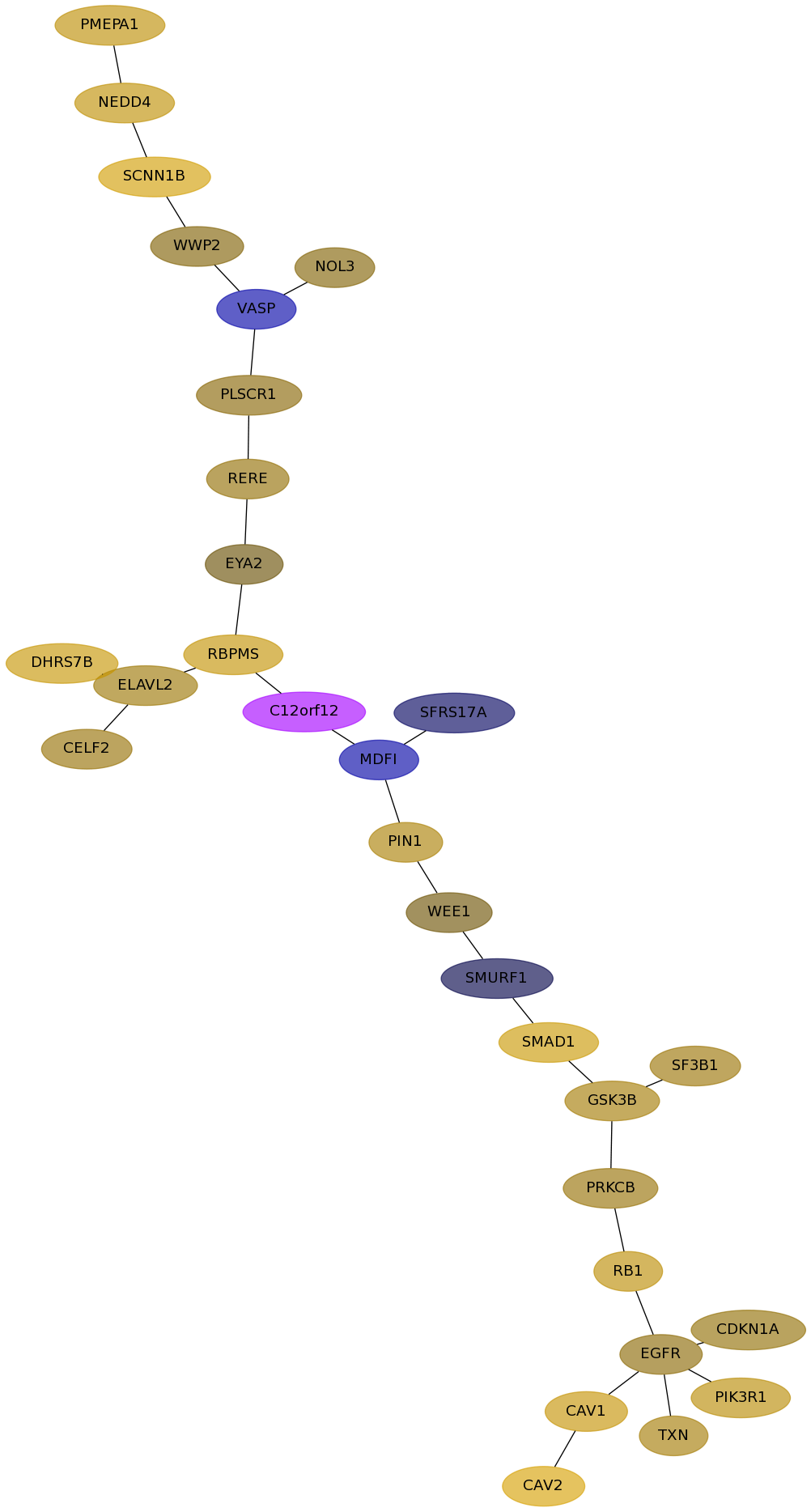
Score for each gene in subnetwork 1317 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| CELF2 |   | 3 | 265 | 634 | 620 | 0.083 | -0.124 | -0.086 | -0.056 | 0.002 |
|---|
| TXN |   | 5 | 148 | 657 | 634 | 0.113 | 0.124 | 0.012 | 0.014 | -0.006 |
|---|
| PLSCR1 |   | 7 | 100 | 634 | 614 | 0.058 | 0.202 | -0.159 | 0.091 | -0.105 |
|---|
| SF3B1 |   | 15 | 40 | 148 | 144 | 0.091 | -0.154 | 0.126 | -0.073 | -0.204 |
|---|
| C12orf12 |   | 3 | 265 | 419 | 419 | undef | 0.135 | 0.002 | undef | undef |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SFRS17A |   | 2 | 391 | 634 | 628 | -0.022 | -0.036 | 0.169 | -0.225 | -0.078 |
|---|
| CAV1 |   | 10 | 67 | 439 | 428 | 0.251 | -0.092 | 0.124 | 0.079 | 0.119 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| NOL3 |   | 1 | 652 | 1385 | 1387 | 0.051 | 0.000 | 0.164 | -0.168 | 0.193 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| NEDD4 |   | 2 | 391 | 1385 | 1385 | 0.222 | -0.099 | 0.196 | -0.005 | 0.146 |
|---|
| RERE |   | 4 | 198 | 634 | 615 | 0.077 | -0.284 | 0.209 | -0.027 | 0.074 |
|---|
| WWP2 |   | 1 | 652 | 1385 | 1387 | 0.049 | -0.107 | 0.134 | 0.017 | 0.280 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| WEE1 |   | 1 | 652 | 1385 | 1387 | 0.031 | -0.051 | 0.163 | 0.034 | 0.215 |
|---|
| ELAVL2 |   | 4 | 198 | 419 | 417 | 0.096 | 0.085 | 0.115 | 0.158 | -0.076 |
|---|
| VASP |   | 1 | 652 | 1385 | 1387 | -0.124 | -0.094 | 0.079 | -0.129 | -0.039 |
|---|
| PMEPA1 |   | 4 | 198 | 937 | 910 | 0.208 | -0.085 | 0.137 | 0.167 | 0.102 |
|---|
| PRKCB |   | 8 | 84 | 33 | 35 | 0.081 | -0.065 | 0.001 | -0.148 | -0.035 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| RBPMS |   | 7 | 100 | 419 | 407 | 0.246 | 0.043 | 0.108 | 0.066 | -0.077 |
|---|
| SCNN1B |   | 2 | 391 | 1385 | 1385 | 0.336 | -0.246 | 0.073 | -0.147 | -0.039 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| SMURF1 |   | 9 | 77 | 33 | 34 | -0.013 | -0.222 | 0.236 | -0.049 | -0.006 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| EYA2 |   | 5 | 148 | 419 | 415 | 0.028 | -0.034 | 0.113 | -0.142 | -0.019 |
|---|
| MDFI |   | 2 | 391 | 634 | 628 | -0.118 | -0.075 | -0.001 | 0.177 | -0.112 |
|---|
| DHRS7B |   | 4 | 198 | 419 | 417 | 0.261 | 0.031 | -0.058 | -0.114 | 0.009 |
|---|
GO Enrichment output for subnetwork 1317 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 4.226E-07 | 9.721E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 4.226E-07 | 4.86E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.658E-07 | 5.104E-04 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 8.431E-07 | 4.848E-04 |
|---|
| protein ubiquitination during ubiquitin-dependent protein catabolic process | GO:0042787 |  | 8.431E-07 | 3.878E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 8.431E-07 | 3.232E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 8.431E-07 | 2.77E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 8.431E-07 | 2.424E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 1.472E-06 | 3.761E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.73E-06 | 3.978E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.888E-06 | 3.947E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| melatonin biosynthetic process | GO:0030187 |  | 3.188E-10 | 7.789E-07 |
|---|
| indolalkylamine biosynthetic process | GO:0046219 |  | 9.544E-10 | 1.166E-06 |
|---|
| indolalkylamine metabolic process | GO:0006586 |  | 4.48E-08 | 3.648E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 2.285E-07 | 1.395E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.684E-07 | 1.8E-04 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 4.56E-07 | 1.857E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 7.963E-07 | 2.779E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 7.963E-07 | 2.432E-04 |
|---|
| protein ubiquitination during ubiquitin-dependent protein catabolic process | GO:0042787 |  | 1.271E-06 | 3.451E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 1.271E-06 | 3.106E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 1.271E-06 | 2.824E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| indolalkylamine biosynthetic process | GO:0046219 |  | 3.394E-10 | 8.167E-07 |
|---|
| indolalkylamine metabolic process | GO:0006586 |  | 2.21E-08 | 2.659E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 2.4E-07 | 1.925E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.181E-07 | 1.913E-04 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 4.79E-07 | 2.305E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 4.79E-07 | 1.921E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 8.366E-07 | 2.875E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 8.366E-07 | 2.516E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 8.366E-07 | 2.236E-04 |
|---|
| protein ubiquitination during ubiquitin-dependent protein catabolic process | GO:0042787 |  | 1.336E-06 | 3.214E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.59E-06 | 3.477E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 4.226E-07 | 9.721E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 4.226E-07 | 4.86E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.658E-07 | 5.104E-04 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 8.431E-07 | 4.848E-04 |
|---|
| protein ubiquitination during ubiquitin-dependent protein catabolic process | GO:0042787 |  | 8.431E-07 | 3.878E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 8.431E-07 | 3.232E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 8.431E-07 | 2.77E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 8.431E-07 | 2.424E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 1.472E-06 | 3.761E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.73E-06 | 3.978E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.888E-06 | 3.947E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 4.226E-07 | 9.721E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 4.226E-07 | 4.86E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.658E-07 | 5.104E-04 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 8.431E-07 | 4.848E-04 |
|---|
| protein ubiquitination during ubiquitin-dependent protein catabolic process | GO:0042787 |  | 8.431E-07 | 3.878E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 8.431E-07 | 3.232E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 8.431E-07 | 2.77E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 8.431E-07 | 2.424E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 1.472E-06 | 3.761E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.73E-06 | 3.978E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.888E-06 | 3.947E-04 |
|---|


