Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1280
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3374 | 1.253e-03 | 1.900e-05 | 1.498e-01 | 3.567e-09 |
|---|
| IPC | 0.3150 | 3.910e-01 | 2.170e-01 | 9.442e-01 | 8.011e-02 |
|---|
| Loi | 0.4893 | 3.000e-05 | 0.000e+00 | 4.719e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5948 | 1.000e-06 | 0.000e+00 | 1.237e-01 | 0.000e+00 |
|---|
| Wang | 0.2529 | 9.125e-03 | 7.407e-02 | 2.494e-01 | 1.685e-04 |
|---|
Expression data for subnetwork 1280 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
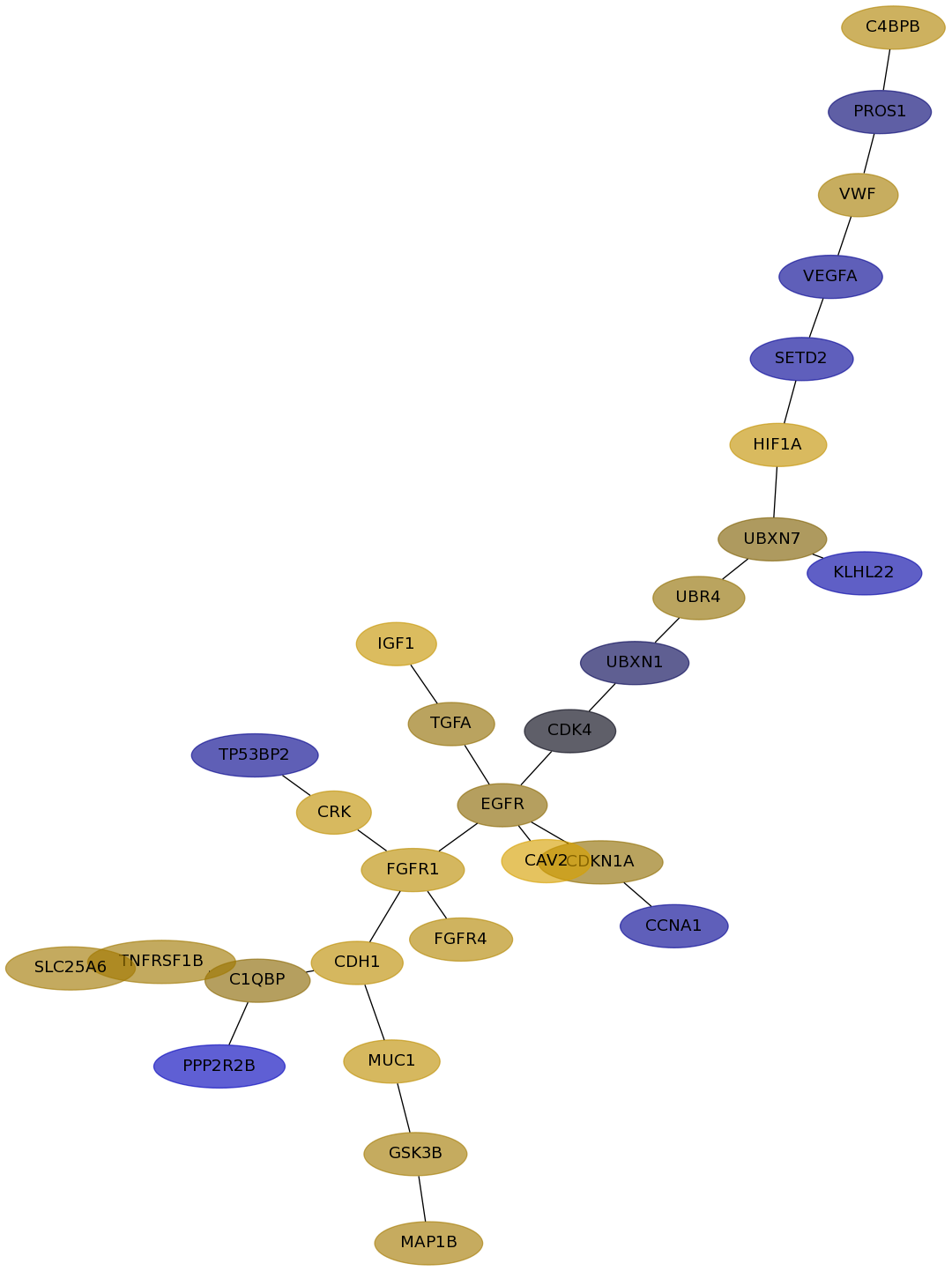
Score for each gene in subnetwork 1280 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| TGFA |   | 8 | 84 | 439 | 429 | 0.077 | -0.057 | -0.027 | -0.158 | 0.020 |
|---|
| CDK4 |   | 1 | 652 | 1220 | 1223 | -0.002 | -0.092 | -0.201 | 0.091 | -0.017 |
|---|
| SETD2 |   | 2 | 391 | 696 | 698 | -0.081 | -0.129 | 0.075 | 0.038 | 0.138 |
|---|
| UBXN1 |   | 1 | 652 | 1220 | 1223 | -0.017 | 0.147 | -0.037 | 0.097 | -0.007 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| C1QBP |   | 4 | 198 | 552 | 547 | 0.064 | 0.134 | -0.072 | 0.080 | -0.006 |
|---|
| UBR4 |   | 2 | 391 | 1039 | 1033 | 0.079 | -0.093 | 0.268 | -0.079 | 0.061 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| C4BPB |   | 1 | 652 | 1220 | 1223 | 0.155 | -0.114 | 0.209 | -0.048 | -0.092 |
|---|
| MAP1B |   | 13 | 47 | 148 | 148 | 0.118 | -0.123 | 0.263 | -0.107 | 0.100 |
|---|
| TNFRSF1B |   | 12 | 55 | 563 | 544 | 0.109 | 0.024 | -0.187 | -0.096 | -0.106 |
|---|
| UBXN7 |   | 6 | 120 | 148 | 156 | 0.049 | -0.055 | 0.153 | 0.224 | 0.175 |
|---|
| KLHL22 |   | 1 | 652 | 1220 | 1223 | -0.121 | 0.087 | -0.010 | 0.081 | 0.109 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| IGF1 |   | 6 | 120 | 398 | 386 | 0.265 | -0.207 | -0.028 | 0.059 | -0.036 |
|---|
| VWF |   | 4 | 198 | 57 | 68 | 0.126 | -0.091 | 0.072 | 0.152 | 0.100 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| PROS1 |   | 1 | 652 | 1220 | 1223 | -0.035 | -0.083 | 0.065 | 0.201 | 0.033 |
|---|
| TP53BP2 |   | 5 | 148 | 1 | 22 | -0.067 | 0.245 | -0.012 | 0.129 | 0.017 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| SLC25A6 |   | 3 | 265 | 778 | 770 | 0.111 | -0.178 | 0.115 | 0.040 | -0.080 |
|---|
GO Enrichment output for subnetwork 1280 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 4.226E-07 | 9.721E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 5.341E-07 | 6.142E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 5.341E-07 | 4.094E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 8.431E-07 | 4.848E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 8.431E-07 | 3.878E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 9.863E-07 | 3.781E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 9.999E-07 | 3.285E-04 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 1.445E-06 | 4.154E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.582E-06 | 4.043E-04 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 2.349E-06 | 5.402E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 3.514E-06 | 7.348E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.013E-07 | 2.476E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.869E-07 | 2.283E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 2.205E-07 | 1.795E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 2.62E-07 | 1.6E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 3.329E-07 | 1.627E-04 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 3.623E-07 | 1.475E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 4.22E-07 | 1.473E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 5.648E-07 | 1.725E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 5.648E-07 | 1.533E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 8.458E-07 | 2.066E-04 |
|---|
| regulation of glycolysis | GO:0006110 |  | 8.458E-07 | 1.879E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.478E-07 | 3.556E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 2.508E-07 | 3.017E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 3.166E-07 | 2.539E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 3.629E-07 | 2.183E-04 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 5.086E-07 | 2.448E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 5.687E-07 | 2.28E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 5.96E-07 | 2.048E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 8.232E-07 | 2.476E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 1.233E-06 | 3.295E-04 |
|---|
| regulation of glycolysis | GO:0006110 |  | 1.233E-06 | 2.966E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 1.233E-06 | 2.696E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 4.226E-07 | 9.721E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 5.341E-07 | 6.142E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 5.341E-07 | 4.094E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 8.431E-07 | 4.848E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 8.431E-07 | 3.878E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 9.863E-07 | 3.781E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 9.999E-07 | 3.285E-04 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 1.445E-06 | 4.154E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.582E-06 | 4.043E-04 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 2.349E-06 | 5.402E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 3.514E-06 | 7.348E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 4.226E-07 | 9.721E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 5.341E-07 | 6.142E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 5.341E-07 | 4.094E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 8.431E-07 | 4.848E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 8.431E-07 | 3.878E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 9.863E-07 | 3.781E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 9.999E-07 | 3.285E-04 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 1.445E-06 | 4.154E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.582E-06 | 4.043E-04 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 2.349E-06 | 5.402E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 3.514E-06 | 7.348E-04 |
|---|


