Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1238
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3341 | 1.456e-03 | 2.500e-05 | 1.599e-01 | 5.820e-09 |
|---|
| IPC | 0.3307 | 3.654e-01 | 1.932e-01 | 9.391e-01 | 6.630e-02 |
|---|
| Loi | 0.4861 | 3.500e-05 | 0.000e+00 | 4.866e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5993 | 1.000e-06 | 0.000e+00 | 1.143e-01 | 0.000e+00 |
|---|
| Wang | 0.2559 | 8.076e-03 | 6.863e-02 | 2.358e-01 | 1.307e-04 |
|---|
Expression data for subnetwork 1238 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
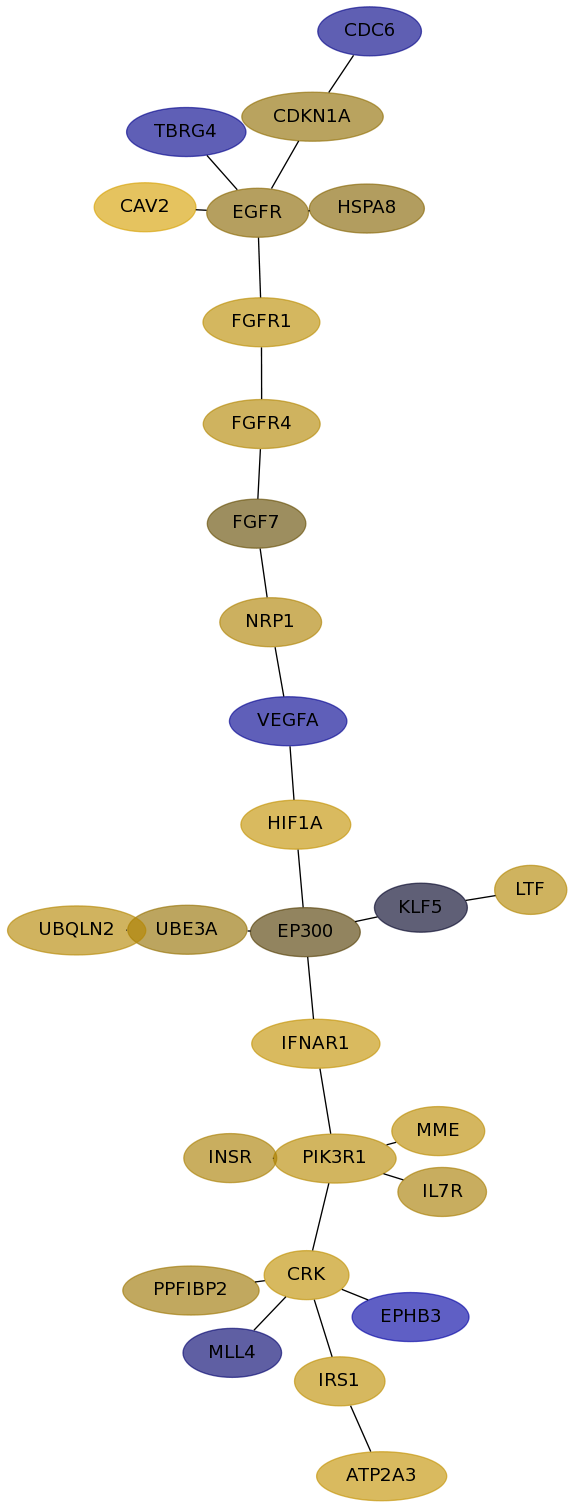
Score for each gene in subnetwork 1238 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| ATP2A3 |   | 9 | 77 | 1 | 18 | 0.240 | -0.040 | 0.082 | -0.229 | -0.025 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| HSPA8 |   | 2 | 391 | 33 | 55 | 0.058 | -0.095 | 0.011 | -0.039 | -0.103 |
|---|
| PPFIBP2 |   | 7 | 100 | 606 | 576 | 0.098 | 0.188 | 0.320 | -0.112 | 0.013 |
|---|
| TBRG4 |   | 2 | 391 | 444 | 458 | -0.066 | 0.173 | -0.060 | -0.073 | -0.055 |
|---|
| CDC6 |   | 4 | 198 | 283 | 289 | -0.059 | 0.198 | -0.053 | 0.038 | 0.095 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| IFNAR1 |   | 1 | 652 | 820 | 835 | 0.244 | -0.121 | 0.067 | 0.188 | -0.004 |
|---|
| UBQLN2 |   | 1 | 652 | 820 | 835 | 0.172 | -0.077 | 0.210 | -0.167 | 0.051 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| LTF |   | 7 | 100 | 492 | 495 | 0.168 | -0.082 | 0.060 | -0.268 | -0.280 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| IRS1 |   | 14 | 42 | 1 | 14 | 0.217 | -0.219 | 0.065 | 0.165 | 0.064 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| UBE3A |   | 2 | 391 | 820 | 815 | 0.085 | -0.043 | -0.018 | 0.183 | 0.017 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| IL7R |   | 14 | 42 | 339 | 334 | 0.130 | -0.151 | -0.112 | -0.130 | -0.160 |
|---|
| MLL4 |   | 4 | 198 | 412 | 409 | -0.032 | -0.093 | 0.052 | -0.101 | -0.153 |
|---|
| FGF7 |   | 8 | 84 | 398 | 384 | 0.026 | -0.094 | 0.237 | 0.070 | 0.083 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| NRP1 |   | 2 | 391 | 536 | 545 | 0.152 | -0.043 | 0.025 | 0.201 | 0.071 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| MME |   | 8 | 84 | 57 | 54 | 0.200 | -0.121 | 0.020 | 0.088 | -0.130 |
|---|
| EPHB3 |   | 4 | 198 | 326 | 333 | -0.115 | -0.048 | 0.135 | 0.099 | -0.008 |
|---|
GO Enrichment output for subnetwork 1238 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.64E-08 | 3.773E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.92E-08 | 3.358E-05 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 3.449E-07 | 2.645E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 5.079E-07 | 2.92E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 6.882E-07 | 3.166E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 6.882E-07 | 2.638E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.103E-06 | 3.624E-04 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 1.103E-06 | 3.171E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.103E-06 | 2.818E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.918E-06 | 4.411E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.918E-06 | 4.01E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell migration | GO:0030335 |  | 4.602E-09 | 1.124E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 6.565E-09 | 8.019E-06 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 9.272E-09 | 7.55E-06 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.179E-07 | 7.203E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.528E-07 | 7.466E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.434E-07 | 1.805E-04 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 4.434E-07 | 1.547E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 6.572E-07 | 2.007E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 6.572E-07 | 1.784E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 6.572E-07 | 1.606E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 9.841E-07 | 2.186E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell migration | GO:0030335 |  | 7.492E-09 | 1.802E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.526E-08 | 1.836E-05 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.601E-07 | 1.284E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.855E-07 | 1.116E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 4.795E-07 | 2.308E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.656E-07 | 2.268E-04 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 5.656E-07 | 1.944E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 8.916E-07 | 2.681E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 8.916E-07 | 2.383E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 1.335E-06 | 3.212E-04 |
|---|
| regulation of glycolysis | GO:0006110 |  | 1.335E-06 | 2.92E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.64E-08 | 3.773E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.92E-08 | 3.358E-05 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 3.449E-07 | 2.645E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 5.079E-07 | 2.92E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 6.882E-07 | 3.166E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 6.882E-07 | 2.638E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.103E-06 | 3.624E-04 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 1.103E-06 | 3.171E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.103E-06 | 2.818E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.918E-06 | 4.411E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.918E-06 | 4.01E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.64E-08 | 3.773E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.92E-08 | 3.358E-05 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 3.449E-07 | 2.645E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 5.079E-07 | 2.92E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 6.882E-07 | 3.166E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 6.882E-07 | 2.638E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.103E-06 | 3.624E-04 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 1.103E-06 | 3.171E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.103E-06 | 2.818E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.918E-06 | 4.411E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.918E-06 | 4.01E-04 |
|---|


