Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1226
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3323 | 1.578e-03 | 2.800e-05 | 1.656e-01 | 7.315e-09 |
|---|
| IPC | 0.3395 | 3.509e-01 | 1.803e-01 | 9.361e-01 | 5.922e-02 |
|---|
| Loi | 0.4849 | 3.700e-05 | 0.000e+00 | 4.925e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6007 | 1.000e-06 | 0.000e+00 | 1.113e-01 | 0.000e+00 |
|---|
| Wang | 0.2569 | 7.778e-03 | 6.704e-02 | 2.318e-01 | 1.209e-04 |
|---|
Expression data for subnetwork 1226 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
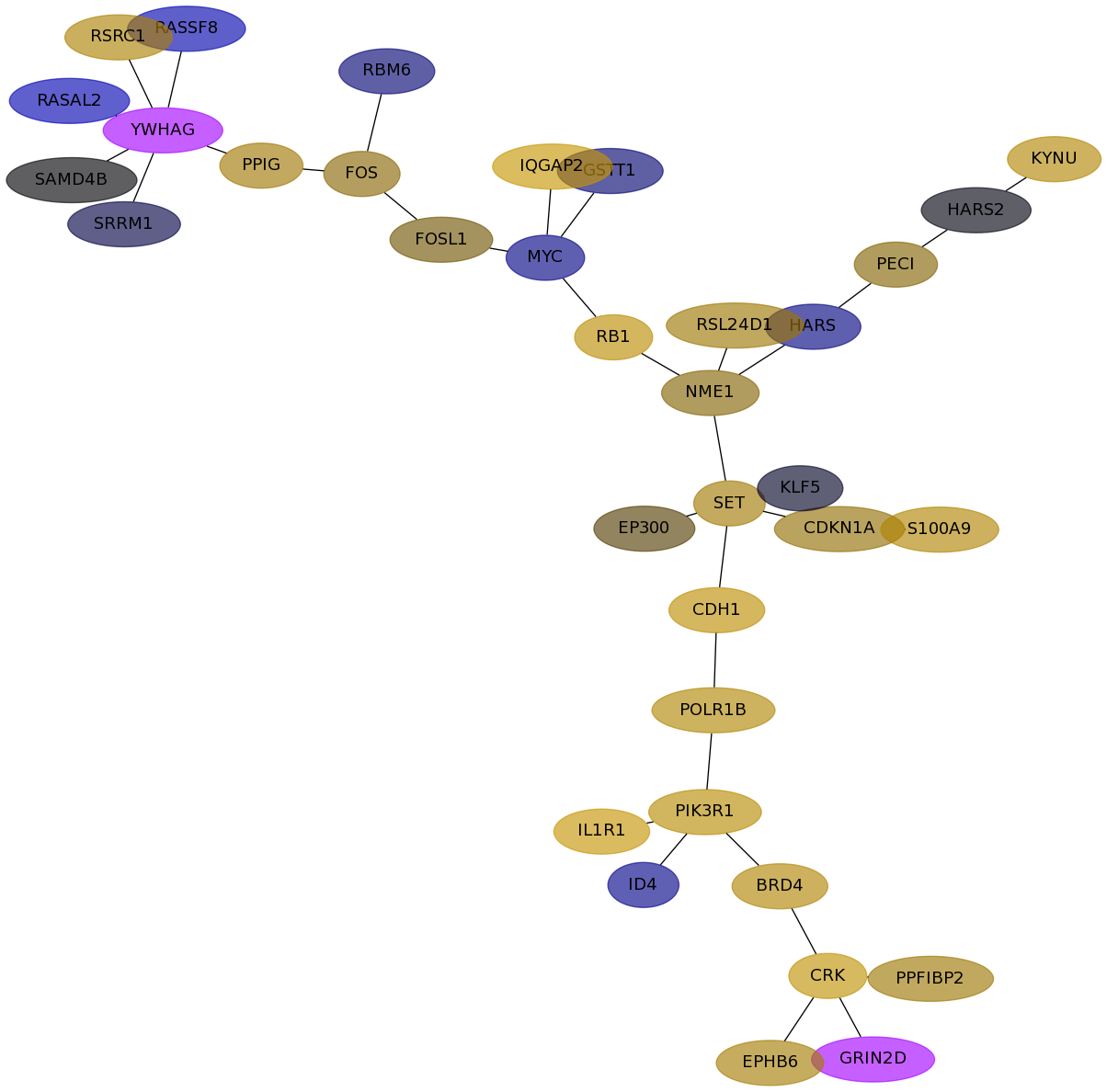
Score for each gene in subnetwork 1226 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| RASAL2 |   | 1 | 652 | 606 | 622 | -0.164 | -0.083 | 0.191 | -0.074 | 0.003 |
|---|
| SRRM1 |   | 1 | 652 | 606 | 622 | -0.012 | -0.120 | 0.139 | 0.244 | -0.109 |
|---|
| PPFIBP2 |   | 7 | 100 | 606 | 576 | 0.098 | 0.188 | 0.320 | -0.112 | 0.013 |
|---|
| GSTT1 |   | 6 | 120 | 364 | 355 | -0.034 | 0.148 | 0.107 | -0.190 | -0.064 |
|---|
| ID4 |   | 13 | 47 | 444 | 430 | -0.060 | -0.179 | 0.137 | 0.241 | 0.036 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| RSRC1 |   | 2 | 391 | 606 | 609 | 0.140 | 0.023 | 0.104 | 0.105 | 0.212 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| RASSF8 |   | 1 | 652 | 606 | 622 | -0.145 | -0.078 | 0.121 | 0.174 | -0.092 |
|---|
| RSL24D1 |   | 2 | 391 | 606 | 609 | 0.098 | 0.206 | -0.026 | 0.169 | -0.022 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| PPIG |   | 3 | 265 | 563 | 556 | 0.107 | -0.116 | 0.024 | 0.221 | 0.101 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| RBM6 |   | 10 | 67 | 148 | 152 | -0.036 | -0.082 | 0.265 | -0.004 | 0.111 |
|---|
| PECI |   | 1 | 652 | 606 | 622 | 0.055 | -0.174 | 0.127 | 0.056 | 0.051 |
|---|
| HARS2 |   | 4 | 198 | 606 | 597 | -0.002 | 0.049 | 0.360 | -0.168 | -0.199 |
|---|
| POLR1B |   | 11 | 59 | 129 | 126 | 0.157 | 0.062 | -0.036 | -0.079 | 0.139 |
|---|
| EPHB6 |   | 7 | 100 | 419 | 407 | 0.122 | 0.008 | 0.042 | -0.219 | -0.059 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| SAMD4B |   | 1 | 652 | 606 | 622 | -0.001 | -0.078 | 0.113 | 0.007 | -0.130 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| HARS |   | 1 | 652 | 606 | 622 | -0.051 | 0.054 | 0.198 | -0.101 | -0.136 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| YWHAG |   | 2 | 391 | 606 | 609 | undef | 0.067 | 0.024 | undef | undef |
|---|
| IL1R1 |   | 8 | 84 | 536 | 515 | 0.257 | -0.211 | 0.178 | -0.071 | 0.003 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| NME1 |   | 7 | 100 | 492 | 495 | 0.054 | 0.267 | 0.106 | 0.046 | -0.035 |
|---|
| KYNU |   | 4 | 198 | 606 | 597 | 0.161 | -0.245 | 0.090 | -0.294 | 0.047 |
|---|
| SET |   | 3 | 265 | 606 | 599 | 0.108 | 0.103 | 0.250 | 0.196 | -0.128 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| FOSL1 |   | 2 | 391 | 606 | 609 | 0.034 | -0.040 | 0.164 | 0.010 | 0.005 |
|---|
GO Enrichment output for subnetwork 1226 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleosome disassembly | GO:0006337 |  | 5.638E-09 | 1.297E-05 |
|---|
| regulation of histone modification | GO:0031056 |  | 1.013E-08 | 1.164E-05 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 1.179E-08 | 9.039E-06 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.814E-07 | 1.043E-04 |
|---|
| cellular macromolecular complex disassembly | GO:0034623 |  | 1.878E-07 | 8.638E-05 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 5.535E-07 | 2.122E-04 |
|---|
| macromolecular complex disassembly | GO:0032984 |  | 5.706E-07 | 1.875E-04 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 9.666E-07 | 2.779E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 1.156E-06 | 2.953E-04 |
|---|
| UTP metabolic process | GO:0046051 |  | 1.543E-06 | 3.549E-04 |
|---|
| CTP metabolic process | GO:0046036 |  | 2.31E-06 | 4.829E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleosome disassembly | GO:0006337 |  | 1.828E-09 | 4.467E-06 |
|---|
| regulation of histone modification | GO:0031056 |  | 3.285E-09 | 4.013E-06 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 4.479E-09 | 3.647E-06 |
|---|
| cellular component disassembly | GO:0022411 |  | 9.146E-08 | 5.586E-05 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 1.179E-07 | 5.763E-05 |
|---|
| cellular macromolecular complex disassembly | GO:0034623 |  | 1.528E-07 | 6.221E-05 |
|---|
| macromolecular complex disassembly | GO:0032984 |  | 3.783E-07 | 1.32E-04 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 4.114E-07 | 1.256E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 5.163E-07 | 1.402E-04 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 6.572E-07 | 1.606E-04 |
|---|
| UTP metabolic process | GO:0046051 |  | 9.841E-07 | 2.186E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleosome disassembly | GO:0006337 |  | 3.204E-09 | 7.709E-06 |
|---|
| regulation of histone modification | GO:0031056 |  | 5.756E-09 | 6.925E-06 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 8.982E-09 | 7.204E-06 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 1.798E-07 | 1.081E-04 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.824E-07 | 8.777E-05 |
|---|
| cellular macromolecular complex disassembly | GO:0034623 |  | 2.669E-07 | 1.07E-04 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 6.268E-07 | 2.155E-04 |
|---|
| macromolecular complex disassembly | GO:0032984 |  | 6.599E-07 | 1.985E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 9.001E-07 | 2.406E-04 |
|---|
| UTP metabolic process | GO:0046051 |  | 1.001E-06 | 2.408E-04 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 1.001E-06 | 2.189E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleosome disassembly | GO:0006337 |  | 5.638E-09 | 1.297E-05 |
|---|
| regulation of histone modification | GO:0031056 |  | 1.013E-08 | 1.164E-05 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 1.179E-08 | 9.039E-06 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.814E-07 | 1.043E-04 |
|---|
| cellular macromolecular complex disassembly | GO:0034623 |  | 1.878E-07 | 8.638E-05 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 5.535E-07 | 2.122E-04 |
|---|
| macromolecular complex disassembly | GO:0032984 |  | 5.706E-07 | 1.875E-04 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 9.666E-07 | 2.779E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 1.156E-06 | 2.953E-04 |
|---|
| UTP metabolic process | GO:0046051 |  | 1.543E-06 | 3.549E-04 |
|---|
| CTP metabolic process | GO:0046036 |  | 2.31E-06 | 4.829E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleosome disassembly | GO:0006337 |  | 5.638E-09 | 1.297E-05 |
|---|
| regulation of histone modification | GO:0031056 |  | 1.013E-08 | 1.164E-05 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 1.179E-08 | 9.039E-06 |
|---|
| cellular component disassembly | GO:0022411 |  | 1.814E-07 | 1.043E-04 |
|---|
| cellular macromolecular complex disassembly | GO:0034623 |  | 1.878E-07 | 8.638E-05 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 5.535E-07 | 2.122E-04 |
|---|
| macromolecular complex disassembly | GO:0032984 |  | 5.706E-07 | 1.875E-04 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 9.666E-07 | 2.779E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 1.156E-06 | 2.953E-04 |
|---|
| UTP metabolic process | GO:0046051 |  | 1.543E-06 | 3.549E-04 |
|---|
| CTP metabolic process | GO:0046036 |  | 2.31E-06 | 4.829E-04 |
|---|


