Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1193
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3315 | 1.634e-03 | 3.000e-05 | 1.681e-01 | 8.239e-09 |
|---|
| IPC | 0.3458 | 3.406e-01 | 1.713e-01 | 9.339e-01 | 5.450e-02 |
|---|
| Loi | 0.4811 | 4.400e-05 | 0.000e+00 | 5.106e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6026 | 1.000e-06 | 0.000e+00 | 1.075e-01 | 0.000e+00 |
|---|
| Wang | 0.2571 | 7.716e-03 | 6.671e-02 | 2.309e-01 | 1.188e-04 |
|---|
Expression data for subnetwork 1193 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
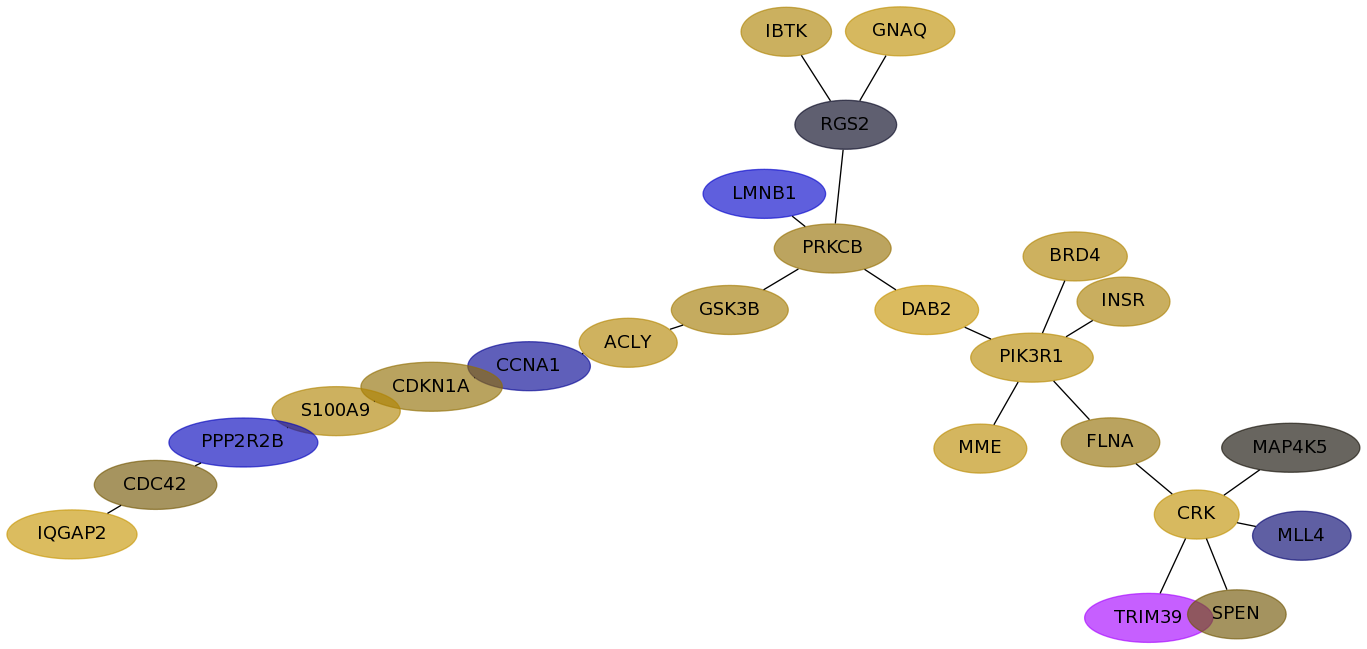
Score for each gene in subnetwork 1193 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| IBTK |   | 1 | 652 | 412 | 446 | 0.146 | 0.233 | 0.261 | -0.102 | 0.236 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| FLNA |   | 7 | 100 | 206 | 198 | 0.077 | 0.081 | 0.107 | 0.148 | 0.126 |
|---|
| RGS2 |   | 2 | 391 | 33 | 55 | -0.003 | 0.118 | 0.061 | 0.084 | 0.178 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| TRIM39 |   | 2 | 391 | 412 | 423 | undef | -0.144 | 0.092 | undef | undef |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| DAB2 |   | 13 | 47 | 1 | 15 | 0.258 | -0.175 | 0.067 | -0.022 | 0.071 |
|---|
| LMNB1 |   | 2 | 391 | 412 | 423 | -0.286 | -0.158 | -0.177 | 0.010 | 0.156 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| MAP4K5 |   | 2 | 391 | 412 | 423 | 0.002 | 0.103 | 0.202 | 0.028 | 0.061 |
|---|
| PRKCB |   | 8 | 84 | 33 | 35 | 0.081 | -0.065 | 0.001 | -0.148 | -0.035 |
|---|
| SPEN |   | 3 | 265 | 129 | 139 | 0.034 | -0.147 | 0.207 | -0.086 | 0.238 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| ACLY |   | 2 | 391 | 412 | 423 | 0.167 | -0.236 | 0.071 | 0.009 | 0.115 |
|---|
| MLL4 |   | 4 | 198 | 412 | 409 | -0.032 | -0.093 | 0.052 | -0.101 | -0.153 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| GNAQ |   | 2 | 391 | 412 | 423 | 0.221 | -0.090 | 0.226 | 0.105 | -0.062 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| MME |   | 8 | 84 | 57 | 54 | 0.200 | -0.121 | 0.020 | 0.088 | -0.130 |
|---|
GO Enrichment output for subnetwork 1193 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.814E-07 | 4.173E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.814E-07 | 2.087E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.622E-07 | 2.777E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.326E-07 | 3.638E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 6.326E-07 | 2.91E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.513E-06 | 5.798E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 2.157E-06 | 7.087E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 2.96E-06 | 8.51E-04 |
|---|
| dopamine receptor signaling pathway | GO:0007212 |  | 2.96E-06 | 7.564E-04 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 2.96E-06 | 6.808E-04 |
|---|
| microspike assembly | GO:0030035 |  | 3.939E-06 | 8.237E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.275E-07 | 3.115E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.275E-07 | 1.558E-04 |
|---|
| interaction with host | GO:0051701 |  | 1.415E-07 | 1.153E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.561E-07 | 2.175E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 3.561E-07 | 1.74E-04 |
|---|
| symbiosis. encompassing mutualism through parasitism | GO:0044403 |  | 5.123E-07 | 2.086E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 5.334E-07 | 1.861E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.045E-06 | 3.191E-04 |
|---|
| dopamine receptor signaling pathway | GO:0007212 |  | 1.391E-06 | 3.777E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.391E-06 | 3.399E-04 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 2.866E-06 | 6.365E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.737E-07 | 4.179E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.737E-07 | 2.089E-04 |
|---|
| interaction with host | GO:0051701 |  | 2.127E-07 | 1.706E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.035E-07 | 1.825E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.035E-07 | 1.46E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 4.849E-07 | 1.944E-04 |
|---|
| symbiosis. encompassing mutualism through parasitism | GO:0044403 |  | 7.69E-07 | 2.643E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.422E-06 | 4.277E-04 |
|---|
| dopamine receptor signaling pathway | GO:0007212 |  | 1.893E-06 | 5.062E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.893E-06 | 4.556E-04 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 3.123E-06 | 6.832E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.814E-07 | 4.173E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.814E-07 | 2.087E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.622E-07 | 2.777E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.326E-07 | 3.638E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 6.326E-07 | 2.91E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.513E-06 | 5.798E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 2.157E-06 | 7.087E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 2.96E-06 | 8.51E-04 |
|---|
| dopamine receptor signaling pathway | GO:0007212 |  | 2.96E-06 | 7.564E-04 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 2.96E-06 | 6.808E-04 |
|---|
| microspike assembly | GO:0030035 |  | 3.939E-06 | 8.237E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.814E-07 | 4.173E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.814E-07 | 2.087E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.622E-07 | 2.777E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.326E-07 | 3.638E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 6.326E-07 | 2.91E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.513E-06 | 5.798E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 2.157E-06 | 7.087E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 2.96E-06 | 8.51E-04 |
|---|
| dopamine receptor signaling pathway | GO:0007212 |  | 2.96E-06 | 7.564E-04 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 2.96E-06 | 6.808E-04 |
|---|
| microspike assembly | GO:0030035 |  | 3.939E-06 | 8.237E-04 |
|---|


