Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1186
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3315 | 1.636e-03 | 3.000e-05 | 1.682e-01 | 8.253e-09 |
|---|
| IPC | 0.3445 | 3.428e-01 | 1.732e-01 | 9.344e-01 | 5.548e-02 |
|---|
| Loi | 0.4800 | 4.600e-05 | 0.000e+00 | 5.163e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6033 | 1.000e-06 | 0.000e+00 | 1.062e-01 | 0.000e+00 |
|---|
| Wang | 0.2570 | 7.736e-03 | 6.681e-02 | 2.312e-01 | 1.195e-04 |
|---|
Expression data for subnetwork 1186 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
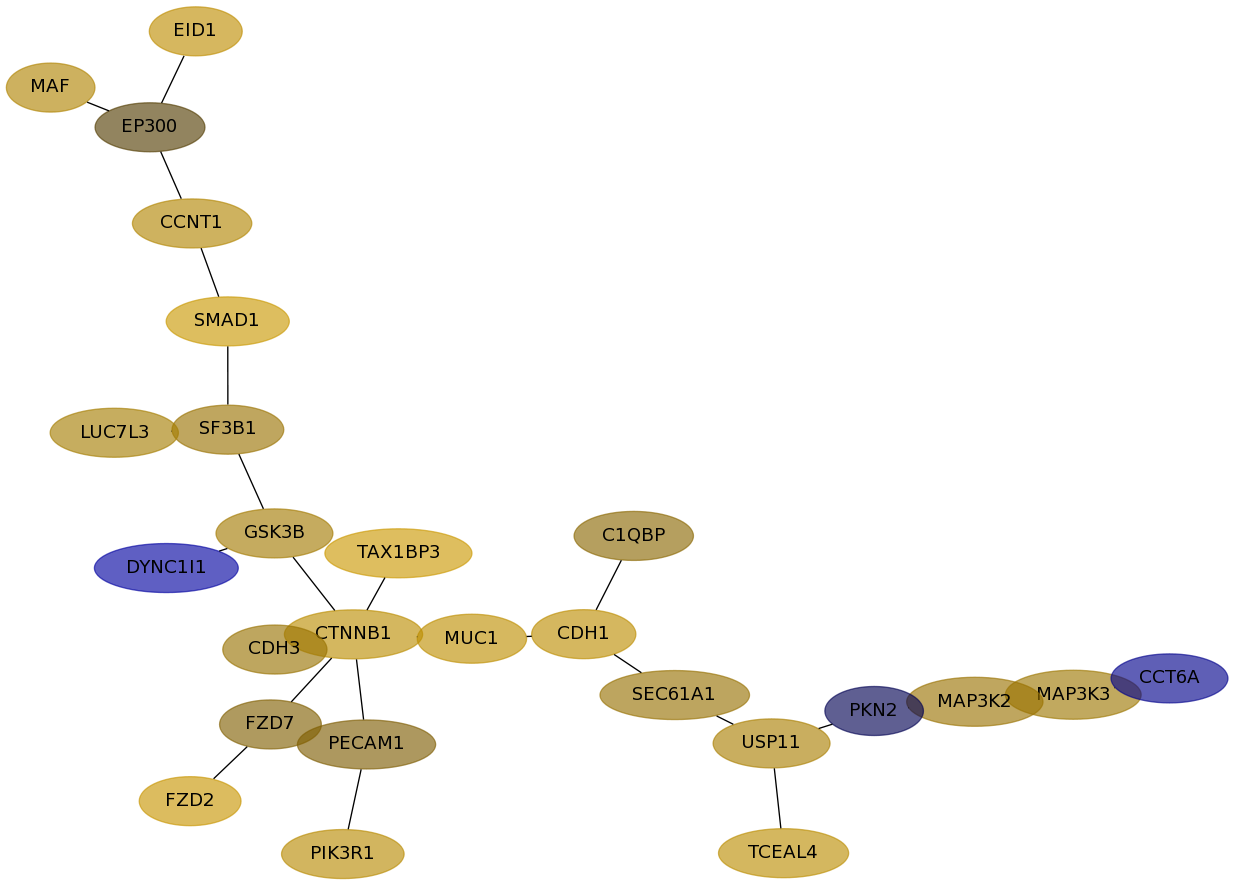
Score for each gene in subnetwork 1186 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| MAP3K3 |   | 4 | 198 | 206 | 205 | 0.102 | 0.085 | 0.046 | 0.028 | 0.094 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| EID1 |   | 4 | 198 | 552 | 547 | 0.212 | -0.069 | 0.051 | -0.010 | -0.037 |
|---|
| SF3B1 |   | 15 | 40 | 148 | 144 | 0.091 | -0.154 | 0.126 | -0.073 | -0.204 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| PECAM1 |   | 3 | 265 | 552 | 550 | 0.046 | 0.014 | -0.161 | 0.040 | -0.106 |
|---|
| FZD2 |   | 8 | 84 | 552 | 540 | 0.273 | -0.067 | -0.081 | -0.067 | 0.081 |
|---|
| DYNC1I1 |   | 5 | 148 | 182 | 181 | -0.109 | -0.069 | 0.013 | -0.086 | 0.088 |
|---|
| C1QBP |   | 4 | 198 | 552 | 547 | 0.064 | 0.134 | -0.072 | 0.080 | -0.006 |
|---|
| FZD7 |   | 5 | 148 | 552 | 541 | 0.051 | -0.219 | 0.062 | 0.062 | -0.059 |
|---|
| PKN2 |   | 3 | 265 | 552 | 550 | -0.017 | -0.198 | 0.158 | 0.060 | 0.068 |
|---|
| MAP3K2 |   | 2 | 391 | 552 | 554 | 0.091 | 0.115 | 0.087 | -0.066 | -0.117 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| SEC61A1 |   | 5 | 148 | 271 | 267 | 0.085 | -0.161 | 0.128 | -0.096 | 0.070 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| LUC7L3 |   | 13 | 47 | 148 | 148 | 0.123 | 0.043 | 0.237 | 0.175 | 0.074 |
|---|
| TCEAL4 |   | 3 | 265 | 552 | 550 | 0.205 | -0.156 | 0.219 | 0.091 | 0.007 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| CCT6A |   | 1 | 652 | 552 | 572 | -0.066 | 0.114 | 0.018 | -0.168 | 0.052 |
|---|
| CDH3 |   | 10 | 67 | 1 | 16 | 0.088 | 0.028 | 0.169 | -0.055 | -0.027 |
|---|
| TAX1BP3 |   | 5 | 148 | 552 | 541 | 0.292 | 0.150 | 0.219 | 0.070 | 0.206 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| USP11 |   | 5 | 148 | 283 | 287 | 0.134 | -0.144 | 0.106 | 0.121 | -0.044 |
|---|
| MAF |   | 2 | 391 | 552 | 554 | 0.155 | 0.024 | 0.085 | -0.094 | 0.122 |
|---|
GO Enrichment output for subnetwork 1186 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.257E-07 | 1.899E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 2.533E-06 | 2.913E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 9.869E-06 | 7.566E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.401E-05 | 8.058E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 9.203E-05 | 0.04233465 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 9.227E-05 | 0.035371 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 9.227E-05 | 0.030318 |
|---|
| establishment of tissue polarity | GO:0007164 |  | 9.227E-05 | 0.02652825 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 9.227E-05 | 0.02358067 |
|---|
| ER overload response | GO:0006983 |  | 9.227E-05 | 0.0212226 |
|---|
| midbrain development | GO:0030901 |  | 9.227E-05 | 0.01929327 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.239E-09 | 5.47E-06 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 1.949E-06 | 2.38E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 4.854E-06 | 3.953E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 6.541E-06 | 3.995E-03 |
|---|
| regulation of chondrocyte differentiation | GO:0032330 |  | 2.7E-05 | 0.01319227 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 3.242E-05 | 0.01320208 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 4.046E-05 | 0.01411947 |
|---|
| mesenchymal cell development | GO:0014031 |  | 4.427E-05 | 0.01352041 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 4.762E-05 | 0.01292735 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.658E-05 | 0.01382229 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 5.658E-05 | 0.01256572 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.232E-09 | 5.37E-06 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 2.727E-06 | 3.281E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 6.788E-06 | 5.444E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 9.145E-06 | 5.501E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 4.162E-05 | 0.02002927 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 5.069E-05 | 0.02032824 |
|---|
| mesenchymal cell development | GO:0014031 |  | 5.729E-05 | 0.01969274 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 6.64E-05 | 0.01996892 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 7.089E-05 | 0.01895036 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 7.089E-05 | 0.01705532 |
|---|
| establishment of tissue polarity | GO:0007164 |  | 7.089E-05 | 0.01550484 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.257E-07 | 1.899E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 2.533E-06 | 2.913E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 9.869E-06 | 7.566E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.401E-05 | 8.058E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 9.203E-05 | 0.04233465 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 9.227E-05 | 0.035371 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 9.227E-05 | 0.030318 |
|---|
| establishment of tissue polarity | GO:0007164 |  | 9.227E-05 | 0.02652825 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 9.227E-05 | 0.02358067 |
|---|
| ER overload response | GO:0006983 |  | 9.227E-05 | 0.0212226 |
|---|
| midbrain development | GO:0030901 |  | 9.227E-05 | 0.01929327 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.257E-07 | 1.899E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 2.533E-06 | 2.913E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 9.869E-06 | 7.566E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.401E-05 | 8.058E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 9.203E-05 | 0.04233465 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 9.227E-05 | 0.035371 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 9.227E-05 | 0.030318 |
|---|
| establishment of tissue polarity | GO:0007164 |  | 9.227E-05 | 0.02652825 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 9.227E-05 | 0.02358067 |
|---|
| ER overload response | GO:0006983 |  | 9.227E-05 | 0.0212226 |
|---|
| midbrain development | GO:0030901 |  | 9.227E-05 | 0.01929327 |
|---|


