Study Desmedt-ER-pos
Study informations
6 subnetworks in total page | file
175 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 176
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.6626 | 1.554e-02 | 3.907e-03 | 8.684e-01 | 5.273e-05 |
|---|
| Loi | 0.4901 | 4.000e-06 | 0.000e+00 | 3.386e-02 | 0.000e+00 |
|---|
| Schmidt | 0.3973 | 4.685e-03 | 5.110e-04 | 8.666e-01 | 2.075e-06 |
|---|
| VanDeVijver | 0.6070 | 1.000e-06 | 2.000e-06 | 3.163e-01 | 6.325e-13 |
|---|
| Wang | 0.2868 | 6.860e-04 | 2.038e-02 | 2.597e-01 | 3.631e-06 |
|---|
Expression data for subnetwork 176 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
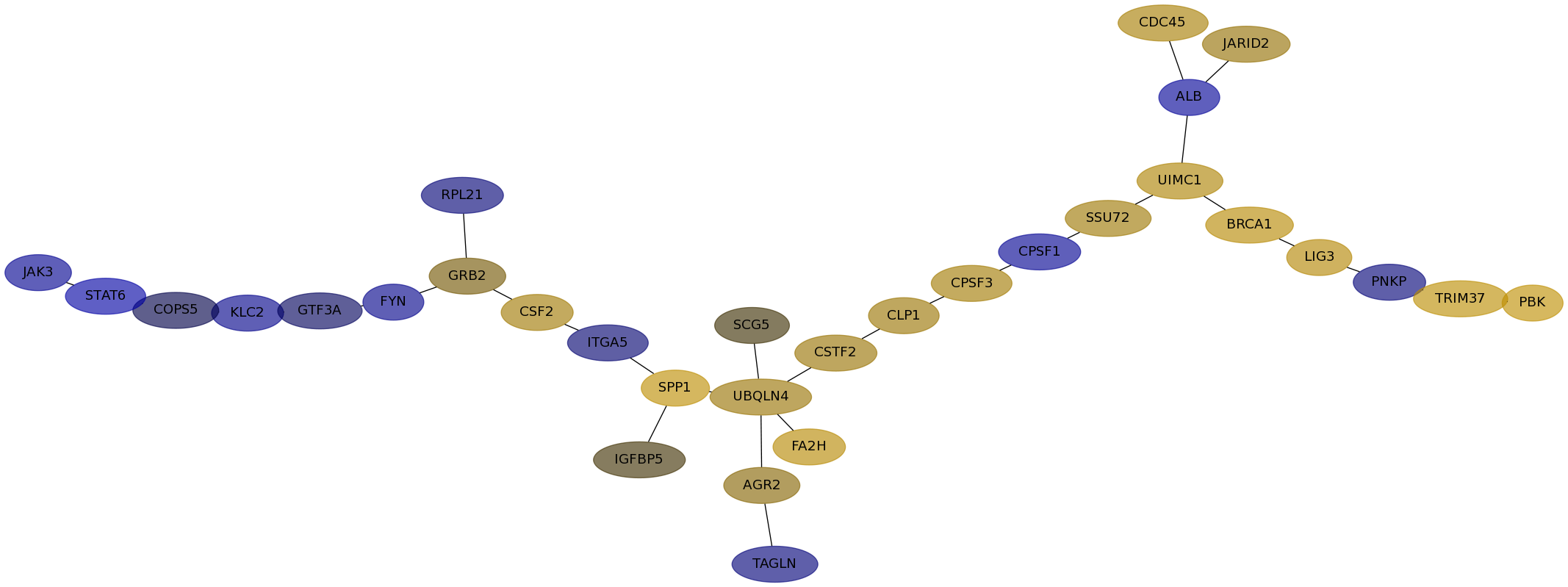
Score for each gene in subnetwork 176 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| UBQLN4 |   | 2 | 9 | 61 | 65 | 0.088 | 0.080 | undef | 0.109 | undef |
|---|
| PBK |   | 1 | 25 | 61 | 72 | 0.216 | 0.198 | 0.166 | 0.092 | 0.267 |
|---|
| RPL21 |   | 1 | 25 | 61 | 72 | -0.039 | -0.003 | -0.163 | -0.073 | -0.169 |
|---|
| KLC2 |   | 1 | 25 | 61 | 72 | -0.064 | 0.124 | 0.027 | undef | 0.223 |
|---|
| AGR2 |   | 2 | 9 | 61 | 65 | 0.056 | 0.223 | 0.057 | undef | 0.034 |
|---|
| CLP1 |   | 1 | 25 | 61 | 72 | 0.093 | 0.042 | 0.118 | -0.085 | 0.065 |
|---|
| ITGA5 |   | 1 | 25 | 61 | 72 | -0.033 | -0.023 | -0.155 | 0.174 | 0.135 |
|---|
| PNKP |   | 1 | 25 | 61 | 72 | -0.040 | 0.231 | -0.001 | 0.268 | 0.101 |
|---|
| GTF3A |   | 1 | 25 | 61 | 72 | -0.021 | 0.172 | 0.033 | undef | -0.017 |
|---|
| BRCA1 |   | 2 | 9 | 61 | 65 | 0.179 | 0.175 | 0.134 | 0.041 | 0.094 |
|---|
| CSF2 |   | 2 | 9 | 61 | 65 | 0.109 | -0.136 | 0.007 | 0.096 | -0.027 |
|---|
| LIG3 |   | 1 | 25 | 61 | 72 | 0.166 | 0.074 | 0.115 | -0.059 | 0.129 |
|---|
| TRIM37 |   | 1 | 25 | 61 | 72 | 0.202 | 0.069 | -0.012 | undef | 0.213 |
|---|
| GRB2 |   | 4 | 1 | 1 | 1 | 0.037 | 0.087 | 0.182 | 0.183 | -0.037 |
|---|
| CDC45 |   | 3 | 3 | 61 | 61 | 0.124 | 0.160 | 0.192 | 0.261 | 0.131 |
|---|
| SPP1 |   | 3 | 3 | 61 | 61 | 0.208 | 0.125 | -0.007 | 0.140 | 0.149 |
|---|
| FYN |   | 1 | 25 | 61 | 72 | -0.064 | -0.062 | -0.113 | 0.162 | -0.085 |
|---|
| CPSF1 |   | 1 | 25 | 61 | 72 | -0.078 | 0.140 | -0.024 | 0.246 | 0.090 |
|---|
| SSU72 |   | 1 | 25 | 61 | 72 | 0.101 | -0.040 | undef | -0.028 | undef |
|---|
| FA2H |   | 1 | 25 | 61 | 72 | 0.181 | 0.050 | 0.207 | undef | 0.128 |
|---|
| ALB |   | 3 | 3 | 61 | 61 | -0.085 | 0.115 | -0.022 | -0.113 | -0.027 |
|---|
| SCG5 |   | 1 | 25 | 61 | 72 | 0.010 | 0.047 | -0.055 | 0.124 | 0.210 |
|---|
| STAT6 |   | 2 | 9 | 45 | 46 | -0.113 | 0.033 | -0.084 | 0.191 | -0.182 |
|---|
| COPS5 |   | 1 | 25 | 61 | 72 | -0.013 | 0.117 | 0.062 | 0.017 | 0.119 |
|---|
| IGFBP5 |   | 3 | 3 | 61 | 61 | 0.011 | 0.158 | 0.087 | 0.185 | -0.000 |
|---|
| CSTF2 |   | 2 | 9 | 61 | 65 | 0.087 | 0.183 | 0.016 | 0.134 | 0.153 |
|---|
| UIMC1 |   | 2 | 9 | 61 | 65 | 0.146 | 0.050 | -0.026 | -0.072 | -0.035 |
|---|
| CPSF3 |   | 1 | 25 | 61 | 72 | 0.112 | 0.111 | undef | -0.052 | undef |
|---|
| JAK3 |   | 2 | 9 | 45 | 46 | -0.075 | 0.070 | -0.100 | 0.171 | 0.081 |
|---|
| JARID2 |   | 2 | 9 | 61 | 65 | 0.079 | 0.005 | 0.116 | 0.116 | 0.004 |
|---|
| TAGLN |   | 1 | 25 | 61 | 72 | -0.042 | -0.047 | -0.184 | 0.113 | -0.007 |
|---|
GO Enrichment output for subnetwork 176 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 5.112E-12 | 1.249E-08 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 1.602E-11 | 1.957E-08 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 2.599E-11 | 2.117E-08 |
|---|
| RNA polyadenylation | GO:0043631 |  | 4.038E-11 | 2.466E-08 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 1.243E-10 | 6.071E-08 |
|---|
| positive regulation of DNA metabolic process | GO:0051054 |  | 8.108E-07 | 3.301E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.152E-06 | 4.021E-04 |
|---|
| peptide hormone processing | GO:0016486 |  | 3.522E-06 | 1.076E-03 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 3.487E-05 | 9.467E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 3.487E-05 | 8.52E-03 |
|---|
| DNA damage response. detection of DNA damage | GO:0042769 |  | 3.487E-05 | 7.745E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 5.075E-12 | 1.221E-08 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 1.59E-11 | 1.913E-08 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 2.581E-11 | 2.07E-08 |
|---|
| RNA polyadenylation | GO:0043631 |  | 4.009E-11 | 2.411E-08 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 1.234E-10 | 5.937E-08 |
|---|
| positive regulation of DNA metabolic process | GO:0051054 |  | 8.11E-07 | 3.252E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.145E-06 | 3.936E-04 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 3.506E-05 | 0.01054563 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 3.506E-05 | 9.374E-03 |
|---|
| DNA damage response. detection of DNA damage | GO:0042769 |  | 3.506E-05 | 8.437E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 5.253E-05 | 0.01149032 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 3.252E-09 | 7.48E-06 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 5.411E-09 | 6.223E-06 |
|---|
| RNA polyadenylation | GO:0043631 |  | 8.489E-09 | 6.508E-06 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 8.489E-09 | 4.881E-06 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 1.271E-08 | 5.847E-06 |
|---|
| positive regulation of DNA metabolic process | GO:0051054 |  | 1.014E-06 | 3.887E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.575E-06 | 5.176E-04 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 5.343E-05 | 0.01535979 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 5.343E-05 | 0.01365314 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 8.002E-05 | 0.01840434 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 8.002E-05 | 0.01673122 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 6.587E-09 | 1.214E-05 |
|---|
| RNA polyadenylation | GO:0043631 |  | 1.183E-08 | 1.09E-05 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 1.967E-08 | 1.209E-05 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 3.085E-08 | 1.421E-05 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 5.037E-07 | 1.857E-04 |
|---|
| positive regulation of DNA metabolic process | GO:0051054 |  | 6.673E-07 | 2.05E-04 |
|---|
| DNA replication | GO:0006260 |  | 1.011E-05 | 2.661E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 1.063E-04 | 0.02448036 |
|---|
| memory | GO:0007613 |  | 1.063E-04 | 0.02176032 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 1.063E-04 | 0.01958429 |
|---|
| interaction with symbiont | GO:0051702 |  | 1.063E-04 | 0.0178039 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 3.252E-09 | 7.48E-06 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 5.411E-09 | 6.223E-06 |
|---|
| RNA polyadenylation | GO:0043631 |  | 8.489E-09 | 6.508E-06 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 8.489E-09 | 4.881E-06 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 1.271E-08 | 5.847E-06 |
|---|
| positive regulation of DNA metabolic process | GO:0051054 |  | 1.014E-06 | 3.887E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.575E-06 | 5.176E-04 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 5.343E-05 | 0.01535979 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 5.343E-05 | 0.01365314 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 8.002E-05 | 0.01840434 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 8.002E-05 | 0.01673122 |
|---|



