Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6629
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7534 | 3.544e-03 | 5.780e-04 | 7.718e-01 | 1.581e-06 |
|---|
| Loi | 0.2304 | 8.011e-02 | 1.132e-02 | 4.278e-01 | 3.880e-04 |
|---|
| Schmidt | 0.6742 | 0.000e+00 | 0.000e+00 | 4.412e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.464e-03 | 0.000e+00 |
|---|
| Wang | 0.2624 | 2.572e-03 | 4.284e-02 | 3.918e-01 | 4.317e-05 |
|---|
Expression data for subnetwork 6629 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
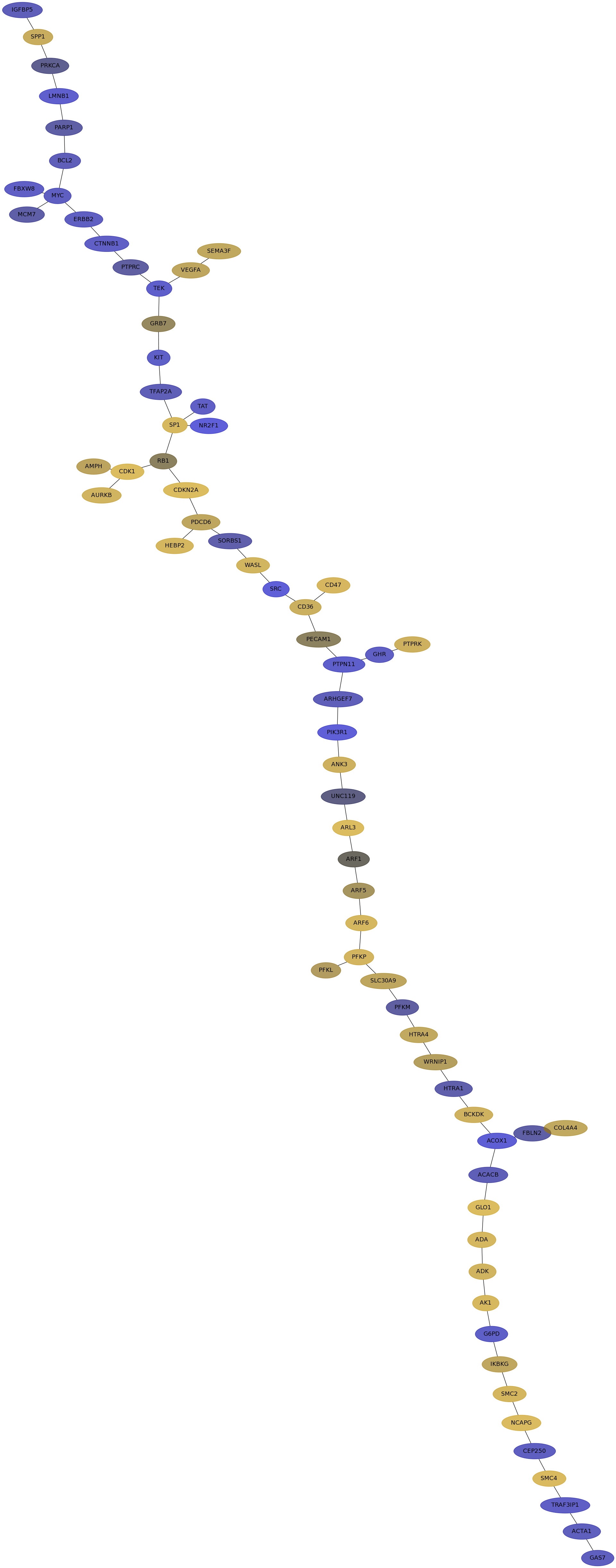
Score for each gene in subnetwork 6629 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| ACACB |   | 2 | 743 | 1 | 66 | -0.069 | 0.025 | 0.127 | -0.130 | -0.006 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| FBLN2 |   | 5 | 360 | 83 | 110 | -0.036 | 0.122 | 0.207 | 0.037 | -0.007 |
|---|
| HTRA1 |   | 7 | 256 | 83 | 102 | -0.046 | 0.184 | 0.051 | 0.132 | 0.224 |
|---|
| ANK3 |   | 6 | 301 | 83 | 105 | 0.153 | 0.034 | 0.001 | 0.094 | -0.038 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| LMNB1 |   | 2 | 743 | 83 | 123 | -0.158 | -0.177 | 0.010 | 0.006 | 0.156 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| PTPRK |   | 14 | 117 | 83 | 96 | 0.156 | 0.141 | -0.094 | 0.048 | -0.024 |
|---|
| FBXW8 |   | 2 | 743 | 83 | 123 | -0.113 | 0.192 | undef | 0.219 | undef |
|---|
| TAT |   | 11 | 148 | 83 | 97 | -0.113 | 0.030 | -0.182 | 0.037 | -0.074 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| SEMA3F |   | 2 | 743 | 83 | 123 | 0.097 | 0.035 | 0.094 | undef | -0.068 |
|---|
| GLO1 |   | 5 | 360 | 83 | 110 | 0.266 | -0.089 | 0.135 | 0.100 | 0.076 |
|---|
| AURKB |   | 4 | 440 | 83 | 112 | 0.179 | -0.189 | 0.086 | -0.058 | -0.028 |
|---|
| SMC2 |   | 4 | 440 | 83 | 112 | 0.202 | -0.108 | 0.119 | 0.132 | 0.079 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| CD36 |   | 2 | 743 | 83 | 123 | 0.146 | 0.017 | 0.129 | 0.061 | 0.124 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| AK1 |   | 1 | 1195 | 83 | 156 | 0.215 | 0.090 | 0.171 | -0.160 | 0.084 |
|---|
| IKBKG |   | 1 | 1195 | 83 | 156 | 0.094 | -0.037 | -0.153 | -0.144 | 0.051 |
|---|
| CEP250 |   | 1 | 1195 | 83 | 156 | -0.098 | 0.014 | -0.241 | 0.109 | 0.145 |
|---|
| WASL |   | 1 | 1195 | 83 | 156 | 0.186 | -0.065 | -0.021 | -0.030 | -0.084 |
|---|
| G6PD |   | 2 | 743 | 83 | 123 | -0.132 | -0.041 | -0.255 | 0.230 | 0.012 |
|---|
| ACTA1 |   | 4 | 440 | 83 | 112 | -0.090 | -0.041 | 0.145 | 0.248 | 0.132 |
|---|
| TRAF3IP1 |   | 9 | 196 | 83 | 99 | -0.113 | -0.063 | -0.088 | 0.276 | undef |
|---|
| HTRA4 |   | 8 | 222 | 83 | 101 | 0.102 | undef | undef | undef | undef |
|---|
| ARF6 |   | 1 | 1195 | 83 | 156 | 0.211 | -0.044 | 0.066 | 0.042 | -0.242 |
|---|
| PFKP |   | 6 | 301 | 83 | 105 | 0.195 | -0.076 | -0.040 | undef | 0.094 |
|---|
| SLC30A9 |   | 2 | 743 | 83 | 123 | 0.089 | -0.011 | 0.026 | -0.104 | 0.050 |
|---|
| BCKDK |   | 3 | 557 | 83 | 117 | 0.166 | -0.004 | -0.012 | -0.031 | -0.001 |
|---|
| GAS7 |   | 3 | 557 | 83 | 117 | -0.093 | -0.082 | 0.172 | 0.120 | 0.064 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| ARF1 |   | 1 | 1195 | 83 | 156 | 0.003 | 0.060 | -0.126 | 0.071 | 0.068 |
|---|
| PECAM1 |   | 4 | 440 | 83 | 112 | 0.014 | -0.161 | 0.040 | -0.105 | -0.106 |
|---|
| MCM7 |   | 2 | 743 | 83 | 123 | -0.038 | -0.091 | 0.134 | undef | 0.056 |
|---|
| NCAPG |   | 3 | 557 | 83 | 117 | 0.253 | -0.183 | 0.065 | undef | 0.020 |
|---|
| SRC |   | 1 | 1195 | 83 | 156 | -0.223 | -0.180 | -0.012 | 0.082 | -0.101 |
|---|
| ADA |   | 4 | 440 | 83 | 112 | 0.208 | -0.164 | -0.042 | -0.102 | 0.219 |
|---|
| WRNIP1 |   | 3 | 557 | 83 | 117 | 0.064 | -0.025 | 0.162 | undef | -0.115 |
|---|
| ARL3 |   | 6 | 301 | 83 | 105 | 0.266 | -0.002 | 0.203 | 0.206 | -0.029 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| UNC119 |   | 10 | 167 | 83 | 98 | -0.009 | -0.324 | -0.099 | 0.094 | 0.071 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| PFKL |   | 7 | 256 | 83 | 102 | 0.056 | 0.106 | -0.173 | 0.160 | -0.158 |
|---|
| AMPH |   | 7 | 256 | 83 | 102 | 0.082 | 0.000 | -0.104 | 0.024 | 0.241 |
|---|
| ARF5 |   | 1 | 1195 | 83 | 156 | 0.038 | -0.049 | -0.091 | 0.062 | 0.000 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| TFAP2A |   | 3 | 557 | 83 | 117 | -0.070 | 0.146 | -0.095 | -0.152 | -0.009 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| SMC4 |   | 3 | 557 | 83 | 117 | 0.247 | -0.111 | 0.214 | 0.072 | 0.094 |
|---|
| COL4A4 |   | 6 | 301 | 83 | 105 | 0.102 | 0.155 | -0.039 | 0.295 | 0.068 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| HEBP2 |   | 2 | 743 | 83 | 123 | 0.207 | -0.044 | -0.153 | -0.038 | 0.013 |
|---|
| PFKM |   | 6 | 301 | 83 | 105 | -0.030 | -0.079 | 0.293 | 0.023 | 0.118 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| PTPRC |   | 9 | 196 | 83 | 99 | -0.031 | -0.204 | -0.050 | -0.179 | -0.068 |
|---|
| PDCD6 |   | 10 | 167 | 1 | 40 | 0.091 | -0.060 | undef | undef | undef |
|---|
| ACOX1 |   | 9 | 196 | 1 | 44 | -0.209 | 0.122 | -0.104 | 0.252 | -0.094 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| CD47 |   | 1 | 1195 | 83 | 156 | 0.214 | 0.027 | -0.007 | -0.176 | -0.057 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| ADK |   | 2 | 743 | 83 | 123 | 0.182 | -0.115 | 0.099 | undef | 0.094 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6629 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| organ growth | GO:0035265 |  | 1.203E-10 | 2.94E-07 |
|---|
| regulation of multicellular organism growth | GO:0040014 |  | 2.875E-08 | 3.511E-05 |
|---|
| negative regulation of transport | GO:0051051 |  | 3.255E-08 | 2.651E-05 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.567E-08 | 2.179E-05 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 3.567E-08 | 1.743E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 6.626E-08 | 2.698E-05 |
|---|
| purine salvage | GO:0043101 |  | 7.102E-08 | 2.479E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 7.102E-08 | 2.169E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.273E-07 | 3.455E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 1.273E-07 | 3.109E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.251E-07 | 4.998E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| organ growth | GO:0035265 |  | 1.228E-10 | 2.954E-07 |
|---|
| regulation of multicellular organism growth | GO:0040014 |  | 2.939E-08 | 3.536E-05 |
|---|
| negative regulation of transport | GO:0051051 |  | 3.03E-08 | 2.43E-05 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.631E-08 | 2.184E-05 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 3.631E-08 | 1.747E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 6.774E-08 | 2.716E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 7.23E-08 | 2.485E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.296E-07 | 3.896E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 1.296E-07 | 3.463E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.296E-07 | 5.525E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.521E-07 | 7.701E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| organ growth | GO:0035265 |  | 4.2E-10 | 9.66E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.407E-08 | 1.618E-05 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 4.196E-08 | 3.217E-05 |
|---|
| regulation of multicellular organism growth | GO:0040014 |  | 7.759E-08 | 4.461E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 9.735E-08 | 4.478E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.936E-07 | 7.421E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 1.936E-07 | 6.361E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.112E-07 | 6.071E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.121E-07 | 1.564E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 8.971E-07 | 2.063E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 9.551E-07 | 1.997E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of multicellular organism growth | GO:0040014 |  | 4.561E-07 | 8.406E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.898E-06 | 1.749E-03 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 4.101E-06 | 2.52E-03 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 4.521E-06 | 2.083E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 4.521E-06 | 1.667E-03 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 4.788E-06 | 1.471E-03 |
|---|
| liver development | GO:0001889 |  | 7.48E-06 | 1.97E-03 |
|---|
| focal adhesion formation | GO:0048041 |  | 8.992E-06 | 2.072E-03 |
|---|
| ruffle organization | GO:0031529 |  | 8.992E-06 | 1.841E-03 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 1.114E-05 | 2.053E-03 |
|---|
| regulation of receptor-mediated endocytosis | GO:0048259 |  | 1.565E-05 | 2.622E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| organ growth | GO:0035265 |  | 4.2E-10 | 9.66E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.407E-08 | 1.618E-05 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 4.196E-08 | 3.217E-05 |
|---|
| regulation of multicellular organism growth | GO:0040014 |  | 7.759E-08 | 4.461E-05 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 9.735E-08 | 4.478E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.936E-07 | 7.421E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 1.936E-07 | 6.361E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.112E-07 | 6.071E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.121E-07 | 1.564E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 8.971E-07 | 2.063E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 9.551E-07 | 1.997E-04 |
|---|



