Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 9775-11-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2128 | 9.645e-03 | 7.190e-03 | 8.472e-02 |
|---|
| IPC-NIBC-129 | 0.2250 | 4.437e-02 | 6.420e-02 | 4.123e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2516 | 1.107e-02 | 1.825e-02 | 2.879e-01 |
|---|
| Loi_GPL570 | 0.3057 | 2.637e-02 | 3.050e-02 | 1.289e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1629 | 1.700e-05 | 1.370e-04 | 1.440e-04 |
|---|
| Parker_GPL1390 | 0.2351 | 1.243e-01 | 1.475e-01 | 6.188e-01 |
|---|
| Parker_GPL887 | 0.0535 | 3.324e-01 | 3.509e-01 | 3.584e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3757 | 7.827e-01 | 7.374e-01 | 9.983e-01 |
|---|
| Schmidt | 0.1785 | 1.815e-02 | 2.086e-02 | 2.189e-01 |
|---|
| Sotiriou | 0.2926 | 1.679e-02 | 1.305e-02 | 2.517e-01 |
|---|
| Van-De-Vijver | 0.2656 | 7.280e-04 | 1.047e-03 | 4.736e-02 |
|---|
| Zhang | 0.2766 | 1.285e-01 | 8.337e-02 | 2.708e-01 |
|---|
| Zhou | 0.4381 | 5.756e-02 | 4.547e-02 | 3.514e-01 |
|---|
Expression data for subnetwork 9775-11-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
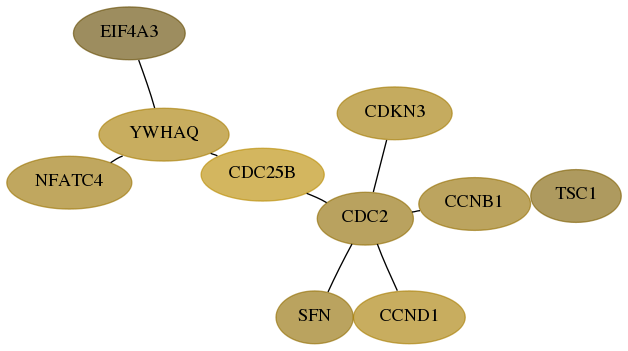
Score for each gene in subnetwork 9775-11-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| nfatc4 |   | 15 | 11 | 1 | 5 | 0.094 | -0.056 | -0.021 | 0.204 | 0.079 | -0.008 | 0.243 | 0.060 | 0.077 | -0.012 | 0.136 | 0.131 | 0.154 |
|---|
| eif4a3 |   | 1 | 116 | 183 | 189 | 0.025 | 0.113 | 0.123 | 0.060 | 0.113 | 0.238 | 0.008 | 0.181 | 0.106 | 0.038 | 0.137 | 0.014 | 0.213 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| ywhaq |   | 5 | 40 | 1 | 9 | 0.129 | 0.110 | 0.049 | 0.116 | 0.071 | 0.160 | -0.210 | 0.038 | 0.007 | 0.054 | 0.072 | 0.138 | 0.187 |
|---|
| cdc25b |   | 4 | 50 | 140 | 128 | 0.197 | 0.107 | 0.134 | 0.223 | 0.092 | 0.194 | 0.071 | 0.200 | 0.067 | 0.115 | undef | 0.048 | 0.153 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
GO Enrichment output for subnetwork 9775-11-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 1.969E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 2.843E-07 | 3.269E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.149E-07 | 5.481E-04 |
|---|
| interphase | GO:0051325 |  | 7.842E-07 | 4.509E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 2.842E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 4.138E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.679E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 4.106E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.875E-05 | 3.92E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.262E-08 | 5.525E-05 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.468E-07 | 1.793E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.04E-07 | 1.661E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.937E-06 | 1.183E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.705E-06 | 1.321E-03 |
|---|
| female meiosis | GO:0007143 |  | 2.705E-06 | 1.101E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 4.055E-06 | 1.415E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.348E-06 | 1.939E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.635E-06 | 1.801E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 7.545E-06 | 1.843E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 7.545E-06 | 1.676E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.458E-08 | 8.32E-05 |
|---|
| negative regulation of translation | GO:0017148 |  | 2.03E-07 | 2.443E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.748E-07 | 2.204E-04 |
|---|
| interphase | GO:0051325 |  | 3.114E-07 | 1.873E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.678E-06 | 1.288E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.37E-06 | 1.351E-03 |
|---|
| female meiosis | GO:0007143 |  | 3.37E-06 | 1.158E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.37E-06 | 1.014E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 8.768E-06 | 2.344E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 9.163E-06 | 2.205E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.426E-06 | 2.062E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.262E-08 | 5.525E-05 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.468E-07 | 1.793E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.04E-07 | 1.661E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.937E-06 | 1.183E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.705E-06 | 1.321E-03 |
|---|
| female meiosis | GO:0007143 |  | 2.705E-06 | 1.101E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 4.055E-06 | 1.415E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.348E-06 | 1.939E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.635E-06 | 1.801E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 7.545E-06 | 1.843E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 7.545E-06 | 1.676E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.458E-08 | 8.32E-05 |
|---|
| negative regulation of translation | GO:0017148 |  | 2.03E-07 | 2.443E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.748E-07 | 2.204E-04 |
|---|
| interphase | GO:0051325 |  | 3.114E-07 | 1.873E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.678E-06 | 1.288E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.37E-06 | 1.351E-03 |
|---|
| female meiosis | GO:0007143 |  | 3.37E-06 | 1.158E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.37E-06 | 1.014E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 8.768E-06 | 2.344E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 9.163E-06 | 2.205E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.426E-06 | 2.062E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.46E-08 | 5.213E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.099E-07 | 1.963E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.28E-07 | 1.524E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.361E-06 | 2.108E-03 |
|---|
| protein N-terminus binding | GO:0047485 |  | 2.689E-06 | 1.92E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.314E-06 | 1.972E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.969E-06 | 2.535E-03 |
|---|
| poly(A) RNA binding | GO:0008143 |  | 4.969E-06 | 2.218E-03 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 6.445E-06 | 2.557E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 6.954E-06 | 2.483E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.954E-06 | 2.258E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.014E-08 | 3.698E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.696E-08 | 1.221E-04 |
|---|
| interphase | GO:0051325 |  | 7.392E-08 | 8.989E-05 |
|---|
| negative regulation of translation | GO:0017148 |  | 8.92E-08 | 8.135E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.34E-06 | 9.773E-04 |
|---|
| protein N-terminus binding | GO:0047485 |  | 1.875E-06 | 1.14E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 2.566E-06 | 1.337E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 2.566E-06 | 1.17E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.848E-06 | 1.56E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.848E-06 | 1.404E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.848E-06 | 1.276E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.458E-08 | 8.32E-05 |
|---|
| negative regulation of translation | GO:0017148 |  | 2.03E-07 | 2.443E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.748E-07 | 2.204E-04 |
|---|
| interphase | GO:0051325 |  | 3.114E-07 | 1.873E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.678E-06 | 1.288E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.37E-06 | 1.351E-03 |
|---|
| female meiosis | GO:0007143 |  | 3.37E-06 | 1.158E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.37E-06 | 1.014E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 8.768E-06 | 2.344E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 9.163E-06 | 2.205E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.426E-06 | 2.062E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 1.969E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 2.843E-07 | 3.269E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.149E-07 | 5.481E-04 |
|---|
| interphase | GO:0051325 |  | 7.842E-07 | 4.509E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 2.842E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 4.138E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.679E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 4.106E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.875E-05 | 3.92E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 1.969E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 2.843E-07 | 3.269E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.149E-07 | 5.481E-04 |
|---|
| interphase | GO:0051325 |  | 7.842E-07 | 4.509E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 2.842E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 4.138E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.679E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 4.106E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.875E-05 | 3.92E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 8.086E-08 | 1.49E-04 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 9.493E-06 | 8.748E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 9.493E-06 | 5.832E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 9.493E-06 | 4.374E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 9.493E-06 | 3.499E-03 |
|---|
| negative regulation of cellular protein metabolic process | GO:0032269 |  | 1.238E-05 | 3.803E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 1.329E-05 | 3.498E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.329E-05 | 3.061E-03 |
|---|
| negative regulation of protein metabolic process | GO:0051248 |  | 1.652E-05 | 3.384E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 1.771E-05 | 3.263E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.771E-05 | 2.967E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 1.969E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 2.843E-07 | 3.269E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.149E-07 | 5.481E-04 |
|---|
| interphase | GO:0051325 |  | 7.842E-07 | 4.509E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 2.842E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 4.138E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.679E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 4.106E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.875E-05 | 3.92E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.987E-08 | 1.895E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 4.83E-07 | 5.73E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.972E-07 | 4.724E-04 |
|---|
| interphase | GO:0051325 |  | 6.545E-07 | 3.883E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.545E-06 | 2.632E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.06E-06 | 2.397E-03 |
|---|
| female meiosis | GO:0007143 |  | 6.06E-06 | 2.054E-03 |
|---|
| oocyte maturation | GO:0001556 |  | 6.06E-06 | 1.797E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 9.085E-06 | 2.395E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.271E-05 | 3.017E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.694E-05 | 3.655E-03 |
|---|



